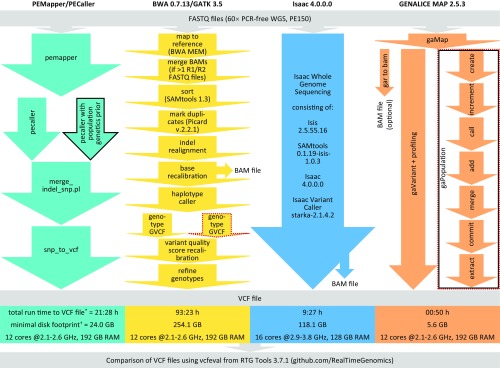
Fig. 1.
Stepwise description, run time, disk footprint, and hardware specifications for the four investigated read mapping and variant calling pipelines. Solid black and dotted red outlines indicate population calling and trio analysis options, respectively. We mapped reads to the GRCh37-like reference genome hs37d5 (8), except for the Isaac pipeline running on BaseSpace Onsite not supporting custom reference genomes, where GRCh37 was used. Notably, hs37d5 contains noncanonical bases which PEMapper/PECaller (downloaded March 29, 2017) was unable to interpret and which were therefore replaced with Ns for this pipeline. Run times shown are for single-sample analyses of the downloaded NA12878 Genome in a Bottle (GIAB) data (legend of Fig. 2) (*). Minimal disk footprints for variant calling (†) were assessed, and thus for GENALICE MAP the size of the optional BAM file was not counted. Analysis parameters: PEMapper/PECaller according to ref. 1; BWA/GATK 3.5 best practices; Isaac default; GENALICE MAP best practices except for max_cigar_complexity = 18, max_context_call_density = 3, and min_map_quality = 1.