Figure 5.
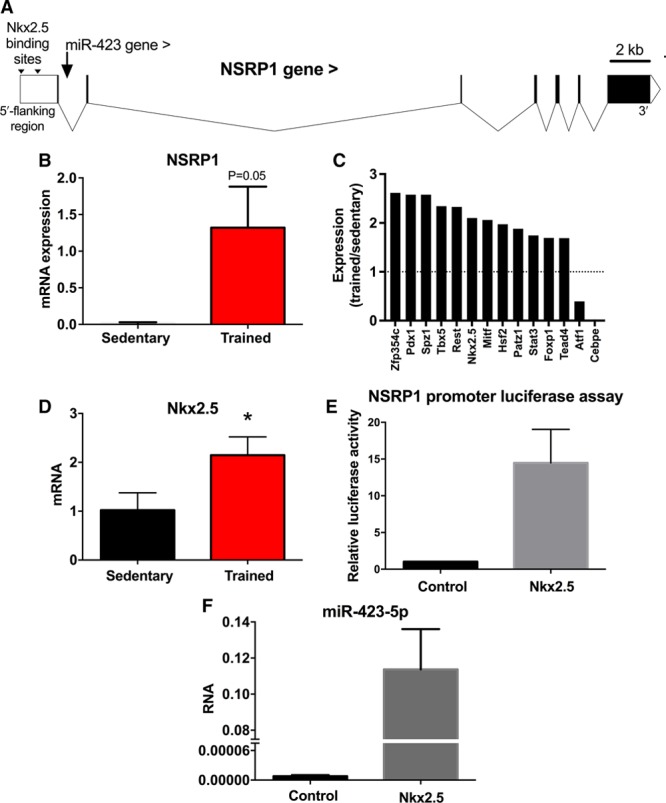
Nkx2.5 regulation of miR-423-5p. A, map of NSRP1 gene (solid blocks show exons with introns in between) showing location of Nkx2.5 binding sites and intronic location of the miR-423 gene. B, Expression of NSRP1 mRNA in sinus node of sedentary and trained mice (n=8/8). C, Significant (P<0.05) training-induced changes in expression of transcription factor transcripts (measured by quantitative real-time reverse transcription polymerase chain reaction [qPCR]) in sinus node of mice. Ratio of mRNA expression in trained mice to expression in sedentary mice shown (n=6/8). D, Expression of Nkx2.5 mRNA in sinus node of sedentary and trained mice (n=8/8). E, Luciferase reporter assay showing activation of NSRP1 transcription by Nkx2.5. H9c2 cells were transfected with 2.1 kb of the 5′ flanking region of NSRP1 cloned into expression vector downstream of gene for luciferase. The cells were cotransfected with Nkx2.5; control cells were not cotransfected with Nkx2.5. Luciferase activity is shown 48 h after transfection. n=3 independent batches of cells with 4 replicates/batch. F, Upregulation of miR-423-5p by Nkx2.5. miR-423-5p expression is shown in H9c2 cells not transfected (control) or transfected with Nkx2.5.
