Figure 6. Neutrophil cPLA2α generates LTB4 production by the neutrophil following infection induced trans-epithelial migration.
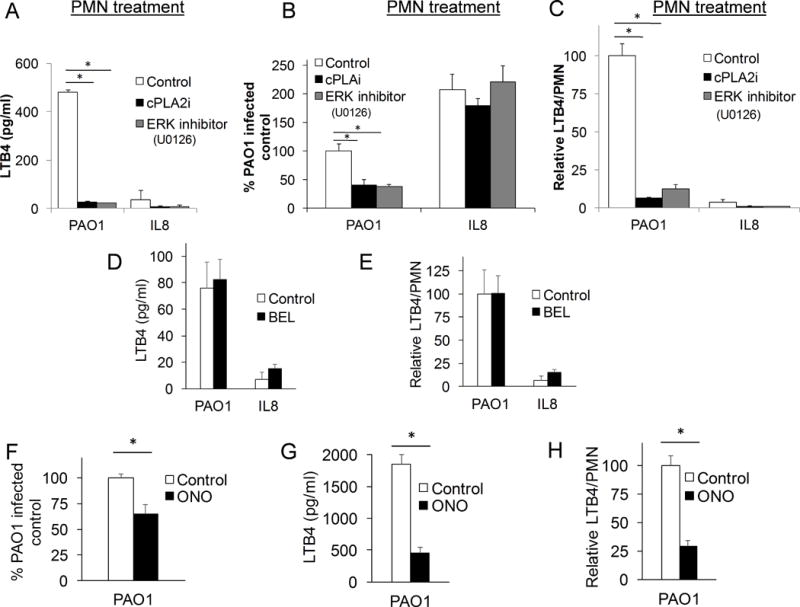
Healthy neutrophils were pretreated for 60 min. with cPLA2α inhibitor (12μM), ERK inhibitor, U0126 (20μM), or vehicle control (DMSO 1:500), then allowed to migrate across an untreated H292 monolayer infected with PAO1 or towards an apical gradient of IL-8. A) After a 2h migration, supernatant was collected from the apical compartment and LTB4 was quantified by ELISA. B) Relative migration was assessed after 2h migration by total myeloperoxidase activity and corrected for by standard curves. C) LTB4 production was adjusted by relative neutrophil migration to calculate relative LTB4 production per neutrophil. Healthy neutrophils were pretreated for 30 min. with iPLA2 inhibitor, BEL (10μM), then allowed to migrate across an untreated H292 monolayer infected with PAO1 or towards an apical gradient of IL-8. D) Supernatant following trans-epithelial migration of BEL-treated PMN was collected from the apical compartment after a 2h migration and LTB4 was quantified by ELISA. E) Relative LTB4 production, calculated as the LTB4/PMN generated as compared to that of PAO1-infected control (see Figure 5B for corresponding migration assay), was assessed. Healthy neutrophils were pretreated for 30 min. with general PLA2 inhibitor, ONO (10μM), then allowed to migrate across an untreated H292 monolayer infected with PAO1. F) Relative migration was assessed after 2h migration by total myeloperoxidase activity and corrected for by standard curves. G) Supernatant following trans-epithelial migration of ONO-treated PMN was collected from the apical compartment after a 2h migration and LTB4 was quantified by ELISA. H) Relative LTB4 production following ONO-treated PMN trans- epithelial migration was assessed. Data are shown as mean corrected value +/− standard deviation and experiments were performed on at least two occasions with n>3 technical replicates. P values involving multiple comparison were calculated by ANOVA with Dunnett’s test (A-C), or when comparing two conditions, p values were calculated by paired Student’s t-test (D-H). P values <0.05 was consider significant.
