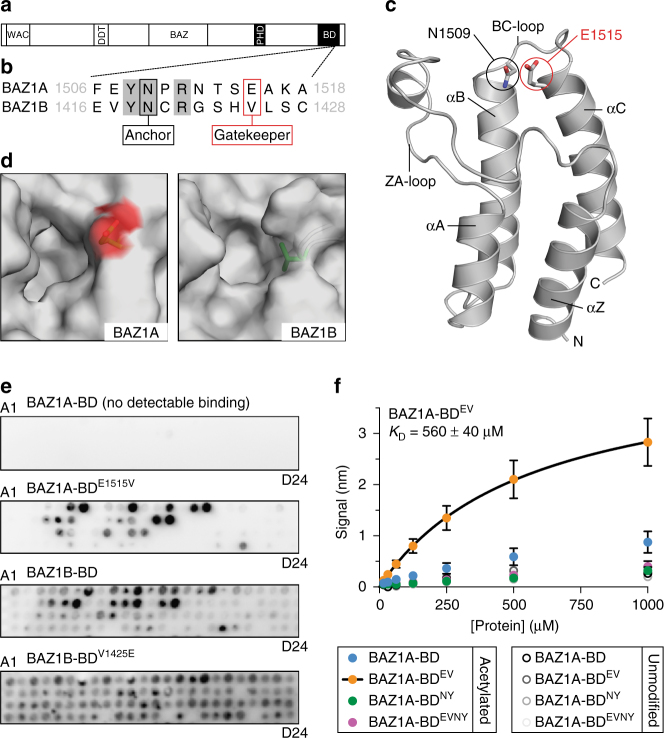
Fig. 3.
A non-canonical glutamic acid “gatekeeper” residue reduces the affinity of BAZ1A-BD for acetylated histone peptides. a Schematic representation of the BAZ1A and BAZ1B proteins. b Sequence of BAZ1A-BD and BAZ1B-BD surrounding the “anchor” and the “gatekeeper” residues; amino-acid boundaries are indicated. c Cartoon representation of BAZ1A-BD from the 1.7 Å resolution crystal structure (Table 1). d Top view of the binding pockets of BAZ1A-BD and BAZ1B-BD (model) in surface representation. The side chains of the gatekeeper residues are shown in stick representation with carbon atoms colored green and the oxygen atoms of the negatively charged BAZ1A-BD E1515 shown in red. e Representative examples of histone peptide array binding images for BAZ1A-BD, BAZ1A-BDE1515V, BAZ1B-BD, and BAZ1B-BDV1425E protein modules. Refer to Supplementary Fig. 3c, d for peptide array information. f Biolayer interferometry binding study of wild-type BAZ1A-BD, BAZ1A-BDE1515V (BDEV), BAZ1A-BDN1509Y (BDNY), or the double mutant BAZ1A-BDE1515V/N1509Y (BDEVNY). Steady-state binding was determined using biotinylated histone H4 1–19, either with or without acetylation of the four lysines present (see “Methods”). Each data point is the mean value ± s.e.m from four independent experiments. The fitted K D for BAZ1A-BDE1515V was determined from the averaged data and reported ± standard error