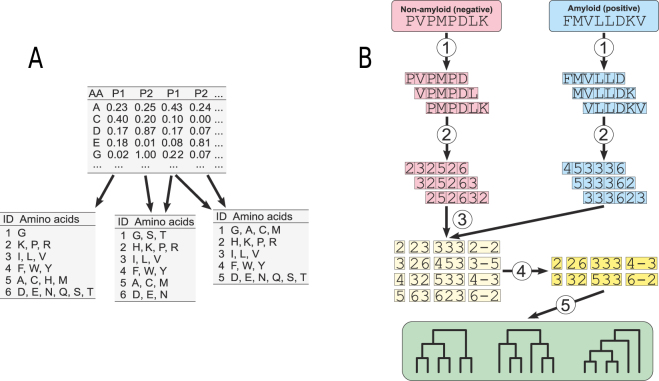
Figure 1.
The scheme of reduced alphabets generation and n-gram extraction from studied peptide sequences. (A) Generation of 18,535 unique amino acid encodings using all possible combinations of selected 17 physicochemical properties. Amino acids (AA) are clustered into groups (ID) using a combination of various physicochemical properties (P1, P2, P3, P4, …). (B) Extraction of n-grams. (1) Extraction of overlapping hexapeptides from peptides with known amyloidicity status. (2) Encoding amino acids of hexapeptides into corresponding groups (reduced alphabet) using alphabets generated (shown in (A)). (3) Extraction of encoded n-grams of different types: continuous with the length from 1 to 3 residues; gapped 2-grams with a gap of the length from 1 to 3 residues; gapped 3-grams with a single gap between residues (not all possibilities are shown). (4) Selection of informative n-grams using Quick Permutation Test (QuiPT). (5) Cross-validation of encodings using random forest classifier, which is trained on the informative n-grams.