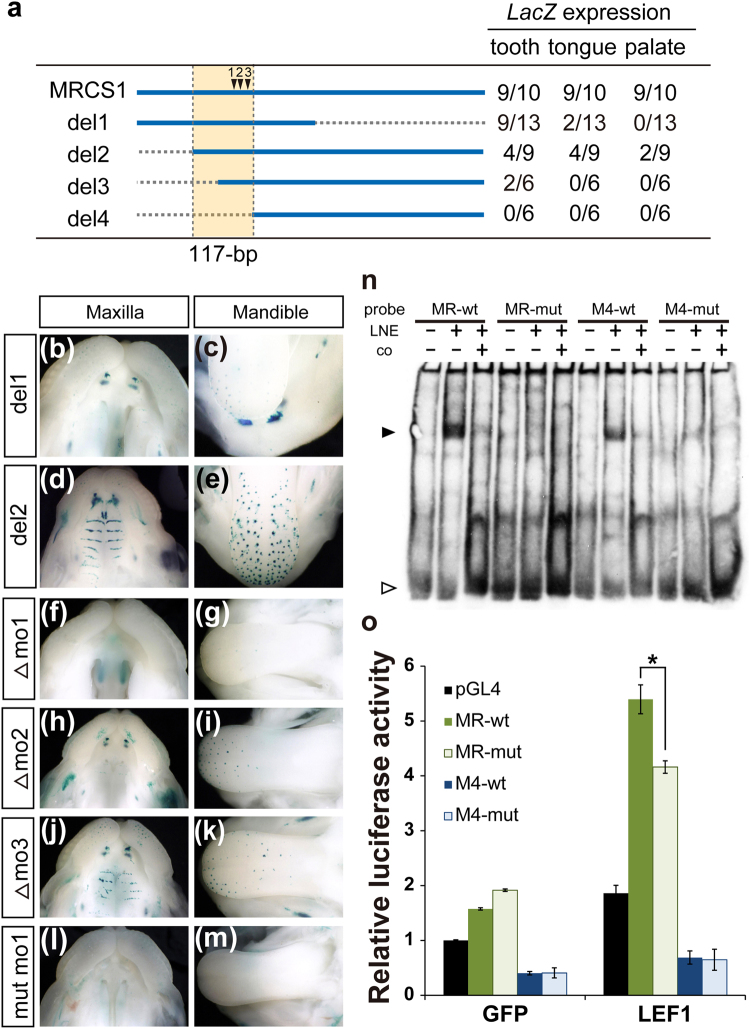
Figure 4.
LacZ expression patterns in transgenic mice with MRCS1 deletion constructs. Diagram of deletion constructs of MRCS1 (a). Dotted lines show the region deleted in each construct. Arrowheads indicate the three motifs shown in Fig. 3. Numbers of LacZ-positive embryos among transgene-positive embryos in each organ are shown in the right column. LacZ expression patterns in the maxilla (b,d) and mandible (c,e) with deletion construct 1 (del1; b,c) and deletion construct 2 (del2; d,e). LacZ expression patterns in transgenic mice with deletions of motif1 (Δmo1: f,g; n = 6), motif2 (Δmo2: h,I; n = 8) and motif3 (Δmo3: j,k; n = 7) and a silent mutation of motif1 (mut mo1: l,m; n = 10). (n) EMSA with sequences at motif1 of MRCS1 and MFCS4 (MR-wt and M4-wt), and sequences with a silent mutation of motif1 (MR-mut and M4-mut, respectively). An arrowhead indicates a specific band shift. An open arrowhead indicates free probes. LNE is an abbreviation of nuclear extracts derived from Lef1-overexpressing cells. Cold oligos as a competitor, co. (o) Relative luciferase activity of the core MRCS1 (MR-wt), the core MRCS1 with a silent mutation of motif1 (MR-mut), the core MFCS4 (M4-wt) and the core MFCS4 with a silent mutation of motif1 (M4-mut) in HEK293T cells. The luciferase reporter constructs containing the core enhancer sequences were cotransfected with plasmid vectors expressing Gfp and Lef1. The reference value for cotransfection with Gfp-expressing and pGL4 empty plasmids was set as 1. Error bars represent the standard deviations obtained from three independent experiments. An asterisk shows a significant difference, as evaluated by Student’s t test (o, p < 0.05).