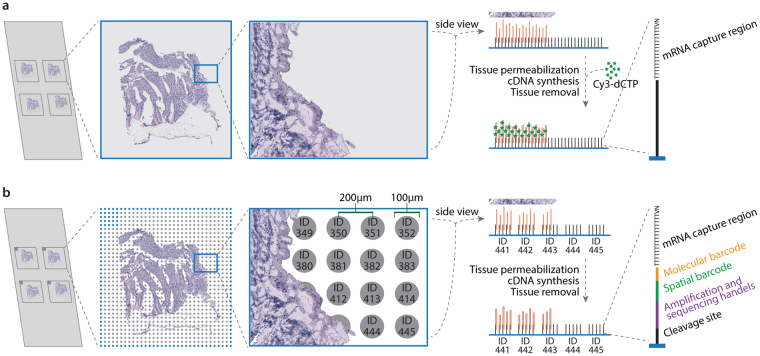
Figure 1.
Schematic of spatially resolved gene expression analysis on adult heart. (a) In order to establish optimal conditions for retrieval and attachment of mRNA in situ, a quality control assay is first used. Intensity of signals generated from the Cy3-cDNA footprint is then scored to examine optimal permeabilization conditions. (b) A spatially barcoded microarray, printed with 1,007 clusters (i.e features) of reverse-transcription oligo-dT primers, are then used with established permebilization conditions. Each feature on the array contains more than 200 million probes, all sharing a unique DNA sequence (barcode) specific to that feature. The barcode is used in the downstream analysis to link each feature’s position within the tissue to the mRNA captured at that position. Finally, following reverse transcription and tissue removal, barcoded cDNA is enzymatically released from the array and used to generate sequencing libraries.