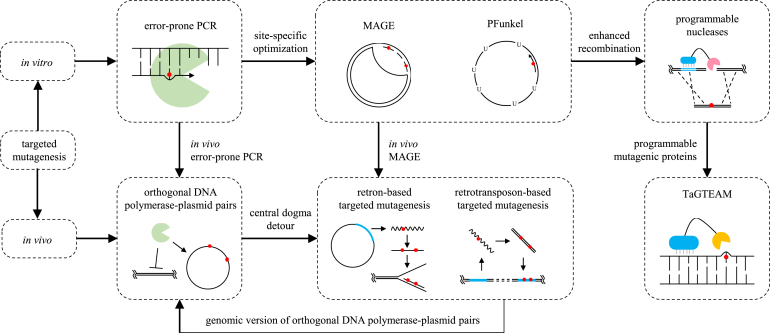
Fig. 1.
Internal logic among existing targeted mutagenesis methods. Targeted mutagenesis methods are classified into the in vitro type and in vivo type. The in vitro type includes error-prone PCR, MAGE, PFunkel and programmable nucleases. Error-prone PCR is adept at discovering key residues within a target gene. Then these residues can be subject to MAGE or PFunkel for further optimization. To compensate for the insufficient recombination level in microbes, programmable nucleases are developed to induce DSBs in the target sites, stimulating the HDR pathway. The in vivo methods take lessons from the in vitro ones. They are mainly orthogonal DNA polymerase-plasmid pairs, retro-element-based targeted mutagenesis and TaGTEAM. Orthogonal DNA polymerase-plasmid pairs are responsible for in vivo error-prone PCR, but there are too few of them to satisfy our need. DNA polymerases directly transfer messages from DNA to DNA, which can also be achieved through the coupling of transcription and reverse transcription processes according to the central dogma. This is what the retro-element-based targeted mutagenesis method is founded on. The E. coli retron-based version of this method is just like an in vivo form of MAGE, while the yeast retrotransposon-based version resembles orthogonal DNA polymerase-plasmid pairs albeit on the genome. As for TaGTEAM, it is actually programmable mutagenic proteins.