ABSTRACT
The Cpx envelope stress response mediates adaptation to stresses that affect protein folding within the envelope of Gram-negative bacteria. Recent transcriptome analyses revealed that the Cpx response impacts genes that affect multiple cellular functions predominantly associated with the cytoplasmic membrane. In this study, we examined the connection between the Cpx response and the respiratory complexes NADH dehydrogenase I and cytochrome bo3 in enteropathogenic Escherichia coli. We found that the Cpx response directly represses the transcription of the nuo and cyo operons and that Cpx-mediated repression of these complexes confers adaptation to stresses that compromise envelope integrity. Furthermore, we found that the activity of the aerobic electron transport chain is reduced in E. coli lacking a functional Cpx response despite no change in the transcription of either the nuo or the cyo operon. Finally, we show that expression of NADH dehydrogenase I and cytochrome bo3 contributes to basal Cpx pathway activity and that overproduction of individual subunits can influence pathway activation. Our results demonstrate that the Cpx response gauges and adjusts the expression, and possibly the function, of inner membrane protein complexes to enable adaptation to envelope stress.
IMPORTANCE Bacterial stress responses allow microbes to survive environmental transitions and conditions, such as those encountered during infection and colonization, that would otherwise kill them. Enteric microbes that inhabit or infect the gut are exposed to a plethora of stresses, including changes in pH, nutrient composition, and the presence of other bacteria and toxic compounds. Bacteria detect and adapt to many of these conditions by using envelope stress responses that measure the presence of stressors in the outermost compartment of the bacterium by monitoring its physiology. The Cpx envelope stress response plays a role in antibiotic resistance and host colonization, and we have shown that it regulates many functions at the bacterial inner membrane. In this report, we describe a novel role for the Cpx response in sensing and controlling the expression of large, multiprotein respiratory complexes at the cytoplasmic membrane of Escherichia coli. The significance of our research is that it will increase our understanding of how these stress responses are involved in antibiotic resistance and the mechanisms used by bacteria to colonize the gut.
KEYWORDS: envelope stress response, NADH dehydrogenase I, cytochrome bo3, membrane protein biogenesis, Cpx envelope stress response, NADH dehydrogenase, cytochrome oxidase, inner membrane, protein complex, protein folding, protein localization, respiration, two-component regulatory systems
INTRODUCTION
Gram-negative bacteria are characterized by the structure of their cell envelope, which consists of the inner membrane (IM), the outer membrane, and the peptidoglycan layer within the periplasmic space. Of these, the IM contains the greatest protein diversity (1). Proteins that reside within the IM play essential roles in energetics, metabolism, transport, and signal transduction. This membrane also serves as a selectively permeable barrier that separates the cytoplasm from the cell's environment. Escherichia coli encodes a suite of envelope stress responses that monitor and maintain envelope integrity, one of which is the Cpx response (2). The Cpx response is controlled by a typical two-component signal transduction system that consists of the membrane-bound sensor kinase CpxA and the cytoplasmic response regulator CpxR. Under inducing conditions, CpxA autophosphorylates at a conserved histidine residue and the phosphate is then transferred to a conserved aspartate residue within CpxR (3). Once phosphorylated, CpxR alleviates envelope stress by altering the transcription of over 100 genes (4–8). In the absence of an inducing cue, CpxA phosphatase activity maintains CpxR in a dephosphorylated and inactive state (3). The auxiliary regulator CpxP inhibits the Cpx response through direct interaction with the sensing domain of CpxA (9, 10).
The Cpx response is thought to detect and respond to potentially lethal misfolded proteins at the bacterial IM. Several conditions predicted or known to generate misfolded IM proteins activate the Cpx response, including overexpression of the outer membrane lipoprotein NlpE, overproduction of pilin subunits in the absence of their cognate chaperones, depletion of the IM protein insertase/assembly factor YidC, mutation of the IM protease FtsH, alkaline pH, and aminoglycoside antibiotics (11–17). Upon induction, CpxR activates the expression of multiple envelope-localized protein-folding and -degrading factors (18–21). Recently, we have shown that the Cpx regulon is enriched for genes encoding IM protein complexes, most of which are downregulated (5).
Complexes of the electron transport chain (ETC) have been identified in all transcriptomic studies of the Cpx response to date (5–8). Enteropathogenic E. coli (EPEC) microarray data indicate that the expression of the genes encoding the respiratory complexes NADH dehydrogenase I (NDH-I) and cytochrome bo3 are among the most strongly downregulated upon activation of the Cpx response (5). NDH-I is one of the entry points for electrons carried by NADH into the bacterial and mitochondrial ETCs. It is one of the largest protein complexes in the E. coli IM, with a molecular mass of 550 kDa (22, 23). It is composed of 13 subunits that are organized into two perpendicular arms, a hydrophobic membrane arm located in the IM and a peripheral arm that protrudes into the cytoplasm (24–26). The subunits of bacterial NDH-I represent the core structure required for the functionality of the human mitochondrial homologue (27, 28). Cytochrome bo3 is a terminal oxidase that couples the oxidation of ubiquinone to the reduction of molecular oxygen. It is composed of four subunits that assemble into a 144-kDa complex within the bacterial IM (29–33). It is a member of the heme-copper oxidase superfamily that also includes cytochrome c oxidase found in human mitochondria (32, 34).
In this study, we tested the hypothesis that the Cpx-mediated downregulation of these large protein complexes is important for adaptation to protein misfolding stresses at the cytoplasmic membrane. We show that the Cpx response regulates the transcription of the genes encoding NDH-I and cytochrome bo3 and further that basal expression of these complexes is sufficient to activate the Cpx response. Intriguingly, our data suggest that Cpx-regulated genes also impact the function, stability, and/or assembly of respiratory complexes, since aerobic respiration is diminished in a ΔcpxRA mutant although transcription is not altered. Cumulatively, our data suggest that the primary function of the Cpx response is to monitor and adjust the biogenesis of macromolecular IM protein complexes.
RESULTS
Regulation of NDH-I and cytochrome bo3 by the Cpx response.
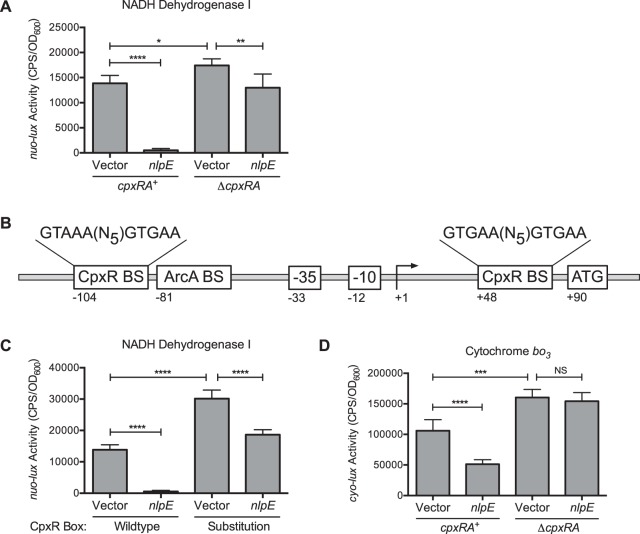
Microarray data indicate that the respiratory complexes NDH-I and cytochrome bo3 are members of the Cpx regulon (5). To confirm these results, we examined the contribution of the Cpx response to the expression of the nuo and cyo gene clusters by using luminescent transcriptional reporters. Activation of the Cpx response by NlpE overexpression resulted in a 26-fold decrease in nuo-lux activity compared to that of the vector control (Fig. 1A). However, when NlpE was overexpressed in a ΔcpxRA mutant, nuo-lux expression was decreased <2-fold (Fig. 1A). These results show that overproduction of NlpE downregulates nuo transcription in a CpxRA-dependent manner. Notably, deletion of cpxRA did not completely abolish the repression of nuo-lux activity upon NlpE overexpression, suggesting that NlpE may regulate this operon through additional signaling pathways. In the absence of stress, there was a small but significant increase in the nuo-lux activity of the ΔcpxRA mutant relative to that of the wild type, suggesting that basal nuo transcription is affected by loss of the Cpx response (Fig. 1A).
FIG 1.
The Cpx response regulates the transcription NDH-I and cytochrome bo3. (A) nuo-lux expression in wild-type and ΔcpxRA mutant EPEC. (B) Schematic representation of the nuo promoter region indicating the locations of the putative CpxR and ArcA binding sites. Numbers indicate distances from the transcription start site in base pairs. −, upstream; +, downstream; BS, binding site. (C) Activity of nuo-lux reporters with a wild-type or mutant CpxR binding site. (D) cyo-lux expression in wild-type and ΔcpxRA mutant EPEC. All luminescence reporters were transformed into EPEC carrying control vector pCA24N or overexpression plasmid pCA-nlpE. Data represent the mean values and standard deviations of five replicate cultures. Asterisks indicate a statistically significant difference from the relevant vector control (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 [one-way ANOVA with Sidak's post hoc test]). NS indicates no statistically significant difference in reporter activity.
A putative CpxR binding site was identified approximately 104 bp upstream of the predicted nuoA transcription start site by using Virtual Footprint (http://prodoric.tu-bs.de/vfp/vfp_promoter.php) (35) (Fig. 1B). To determine if this DNA sequence is required for regulation of nuo transcription by the Cpx response, the putative upstream CpxR binding site (Fig. 1B, bp−104) was mutated from 5′-GTAAA(N5)GTGAA-3′ to 5′-CAGTA(N5)CAGTA-3′ in the nuo-lux reporter. As shown previously, NlpE overexpression strongly reduced activity of the wild-type nuo-lux reporter. However, nuo-lux activity was decreased <2-fold when the putative CpxR binding site was mutated (Fig. 1C). These data support the conclusion that the repression of nuo transcription upon activation of the Cpx response is mediated by the direct binding of CpxR to the nuo promoter region. Interestingly, we observed that basal nuo-lux activity was increased when the putative upstream (Fig. 1B, bp−104) CpxR binding site was mutated in the vector control strain, although the reason for this increase is unknown (Fig. 1C).
In accordance with microarray data, NlpE overexpression resulted in a 2-fold decrease in cyo-lux expression compared to that of the vector control (Fig. 1D). This repression was dependent on the Cpx response, as overexpression of NlpE did not reduce cyo-lux activity in a ΔcpxRA mutant (Fig. 1D). Furthermore, the basal activity of the cyo-lux reporter was slightly increased in the ΔcpxRA mutant in the absence of stress. A putative CpxR binding site was also identified in the cyo promoter, which overlaps the −35 box of the promoter. These observations suggest that CpxR may also bind at the cyo promoter to directly repress transcription.
Cpx pathway activity affects respiration.
Our results suggest that induction of the Cpx pathway would decrease the activity of the aerobic ETC. To confirm our findings, we compared oxygen consumption by wild-type EPEC to that of a cpxA24 mutant, which exhibits constitutive activation of the Cpx response (3). As shown in Fig. 2, the rate of oxygen consumption of the cpxA24 mutant was lower than that of the wild-type strain. As expected, this indicates that decreased expression of respiratory complexes upon activation of the Cpx response leads to decreased activity of the aerobic ETC.
FIG 2.
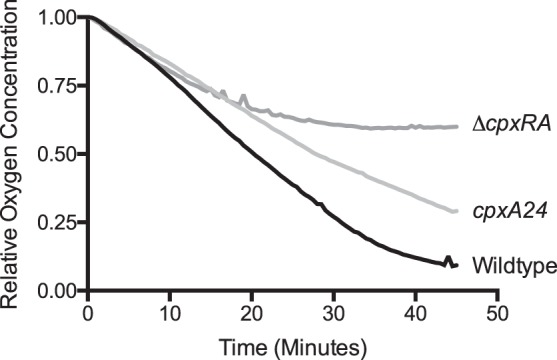
Oxygen consumption is reduced by activation and inhibition of the Cpx response. The oxygen concentration was measured every 30 s for 45 min in a closed system containing wild-type, cpxA24 (Cpx ON), or ΔcpxRA (Cpx OFF) EPEC. The oxygen concentration at each time point was divided by the oxygen concentration at time zero. Data are representative of three independent experiments.
We also compared oxygen consumption by wild-type EPEC to that of a ΔcpxRA mutant to determine the effect of loss of the Cpx response on aerobic respiration. Unexpectedly, oxygen consumption by the ΔcpxRA mutant was lower than that of the wild-type strain (Fig. 2). In replicate experiments, we observed variable rates of respiration in the ΔcpxRA mutant. In some replicates, the initial rate of respiration appeared similar to that of the wild type, but in every instance (five replicates), oxygen consumption slowed relative to the of the wild type over the course of the experiment, and in no case was the ΔcpxRA mutant ever able to consume all of the oxygen in the vial (see Fig. S1 in the supplemental material). This result was surprising, as the transcription of at least the NDH-I and cytochrome bo3 genes is minimally changed in this mutant (Fig. 1). This phenotype was similar to that of a cyo mutant lacking the cytochrome bo3 oxidase (Fig. S1), which indicates that the defect in respiration observed in the ΔcpxRA mutant, at least under these conditions (mid-log-phase cells respiring in terrific broth), is due largely to problems with the biogenesis or function of cytochrome bo3 oxidase. These findings suggest that the Cpx response regulates a factor(s) that facilitates aerobic respiration.
Expression of respiratory complexes is toxic during envelope stress.
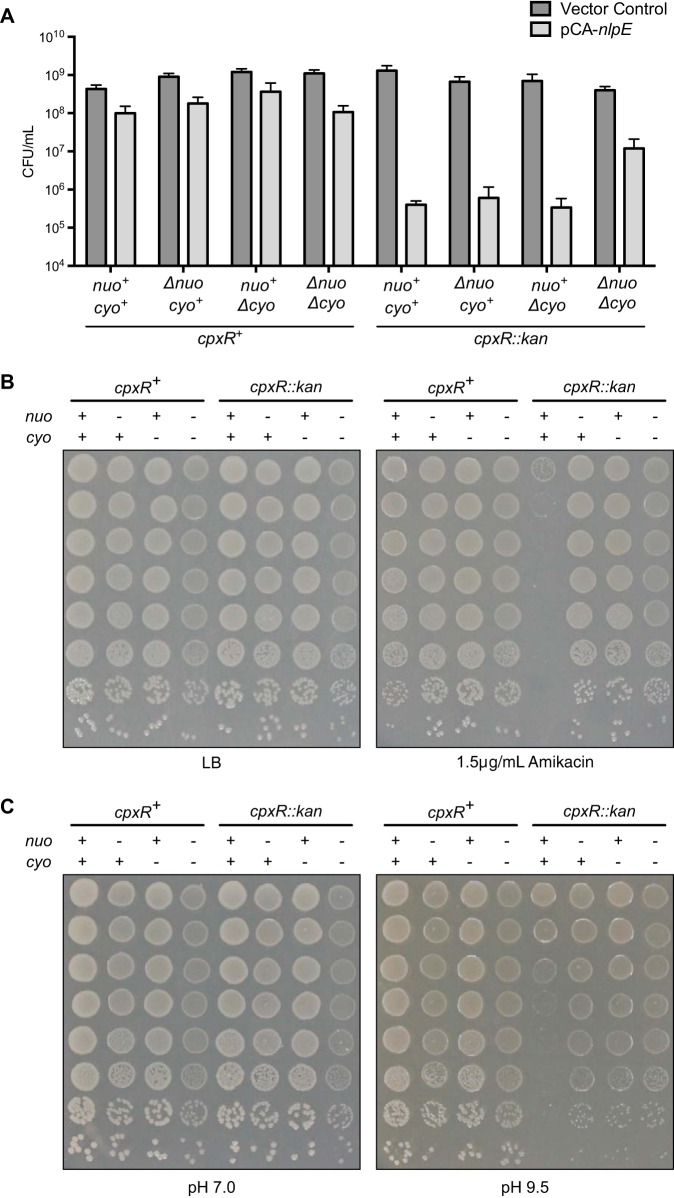
Given the Cpx-mediated downregulation of these large protein complexes, we hypothesized that their presence must be toxic in the presence of envelope stresses. To investigate this possibility, we examined the impact of the NDH-I and cytochrome bo3 oxidase complexes on the toxicity of envelope stresses in a cpxR mutant that is not able to inhibit the expression of the nuo and cyo operons. We first established the sensitivity of wild-type EPEC and its isogenic mutant lacking cpxR to Cpx-sensed envelope stressors previously identified in E. coli K-12, including overproduction of NlpE, aminoglycoside antibiotics, and alkaline pH. NlpE overexpression in the wild-type strain had a mild deleterious effect on growth (Fig. 3A). However, the cpxR mutant was approximately 100-fold more susceptible to the effects of NlpE overexpression than the wild type (Fig. 3A). A similar result was observed when the cpxR mutant was exposed to the aminoglycoside antibiotic amikacin. Wild-type EPEC was resistant to 1.5 μg/ml amikacin; however, the cpxR mutant was susceptible to killing at this concentration (Fig. 3B) and higher concentrations (Fig. S2). It has previously been shown that cpxR is required for E. coli K-12 growth at alkaline pH (16). Here, we confirmed this finding in EPEC, showing that the cpxR mutant has a growth defect at pH 9.5 compared to the wild-type strain (Fig. 3C). Overall, these results show that functional CpxR is required for EPEC to adapt to these envelope stressors.
FIG 3.
Deletion of the nuo and cyo operons in a cpxR mutant restores resistance to envelope stress. (A) IPTG at 1 mM was added to EPEC strains carrying control vector pCA24N or overexpression vector pCA-nlpE to induce overproduction of NlpE. The number of viable bacterial cells present 4.5 h after induction was determined by CFU counting. Data represent the mean values and standard deviations of three replicate cultures. (B and C) Microdilutions of EPEC cultures grown on plain LB or LB supplemented with 1.5 μg/ml amikacin (B) or LB buffered to pH 7.0 or 9.5 (C). Data are representative of at least two independent experiments. The strains shown are as follows: wild-type EPEC; the Δnuo, Δcyo, and cpxR::kan single mutants; the Δnuo Δcyo, Δnuo cpxR::kan, and Δcyo cpxR::kan double mutants; and the Δnuo Δcyo cpxR::kan triple mutant. +, presence of an operon; −, deletion of an operon.
In the absence of CpxR, expression of NDH-I and cytochrome bo3 is not substantially altered in the presence of envelope stress (Fig. 1). To determine if the expression of these complexes contributes to the sensitivity of the cpxR mutant to various stressors, we deleted the nuo and cyo operons in the wild-type and cpxR mutant backgrounds. We then determined if the deletion of these operons could rescue the sensitivity of the cpxR mutant to NlpE overproduction, amikacin, and alkaline pH. The sensitivity of the cpxR mutant to NlpE overexpression was unchanged when either nuo or cyo was individually deleted (Fig. 3A). However, if both nuo and cyo were deleted, the resistance of the cpxR mutant to NlpE overexpression was partially restored to that of EPEC containing a functional Cpx response (Fig. 3A). Further, deletion of nuo, cyo, or both in the cpxR mutant restored resistance to amikacin (Fig. 3B and S2) and growth at alkaline pH (Fig. 3C). Together, these results suggest that the sensitivity of the cpxR mutant to multiple envelope stressors arises, in part, from the inability to downregulate NDH-I and cytochrome bo3 and that the expression of these protein complexes is toxic during envelope stress.
The presence of respiratory complexes contributes to envelope stress sensed by CpxA.
As deletion of nuo and cyo can alleviate the toxicity of certain envelope stresses in a cpxR mutant strain background, we hypothesized that these complexes themselves may generate envelope stress. To examine this possibility, we determined Cpx pathway activity in the Δnuo and Δcyo single mutants and the Δnuo Δcyo double mutant by measuring the activity of a cpxP luminescent transcriptional reporter. cpxP expression is commonly used as a proxy for Cpx pathway activity, as it is one of the most highly transcribed genes upon activation of the Cpx response and its expression depends almost exclusively on CpxR (4, 16, 36). As shown in Fig. 4A, deletion of either nuo or cyo reduced cpxP expression at all stages of growth. The Δnuo Δcyo double mutant had lower cpxP expression than either the Δnuo or the Δcyo single mutant, suggesting an additive effect on Cpx pathway activity. Notably, deletion of these complexes prevented induction of the Cpx response upon entry into stationary phase (Fig. 4A), even though all of the strains were able to grow to stationary phase (Fig. S3). These results suggest that NDH-I and cytochrome bo3 increase basal levels of Cpx pathway activity and may contribute to growth-related activation of the Cpx response.
FIG 4.
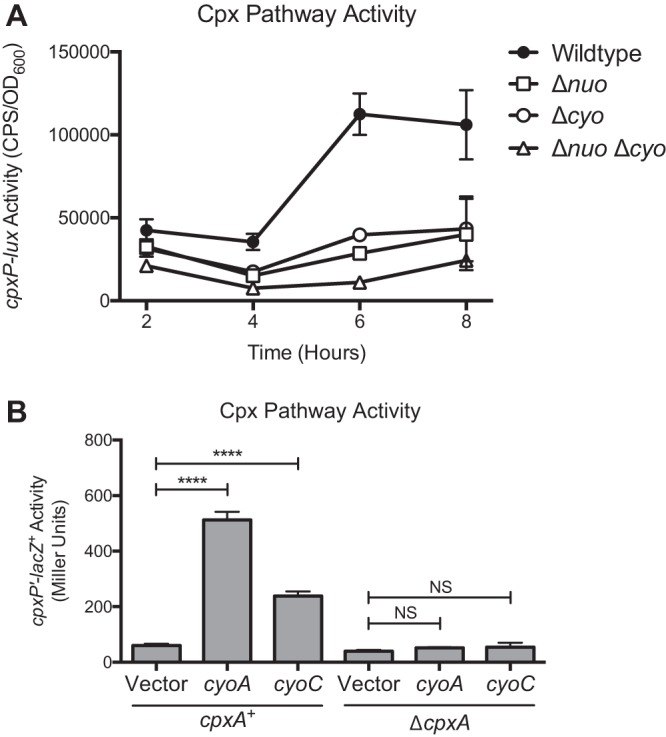
Expression of NDH-I and cytochrome bo3 alters Cpx pathway activity. (A) Time course of cpxP-lux expression in wild-type EPEC, the Δnuo and Δcyo single mutants, and the Δnuo Δcyo double mutant. (B) cpxP-lacZ expression in wild-type and ΔcpxA mutant E. coli MC4100 carrying control vector pCA24N or overexpression vector pCA-cyoA or pCA-cyoC. All data correspond to the means and standard deviations of three replicate cultures. Asterisks indicate a statistically significant difference from the relevant control vector (****, P ≤ 0.0001 [one-way ANOVA with Sidak's post hoc test]). NS indicates no statistically significant difference in reporter activity.
The possibility that the Cpx pathway senses a signal associated with the NDH-I and cytochrome bo3 protein complexes is further supported by results of an independent genetic screen performed to identify proteins that modulate the activity of the Cpx pathway. As shown in Fig. 4B, overexpression of cytochrome bo3 subunits II (CyoA) and III (CyoC) activated the Cpx response approximately 8- and 4-fold, respectively (Fig. 4B). When cpxA was mutated, activation of the Cpx pathway by overexpression of either gene was diminished to <2-fold (Fig. 4B). These data indicate that cytochrome bo3 subunits II and III function as multicopy activators of the Cpx pathway in a CpxA-dependent manner and further reinforce our finding that the Cpx response is sensitive to the presence of the cytochrome bo3 oxidase.
DISCUSSION
Membrane-bound respiratory complexes constitute a major part of the IM proteome (37). Therefore, their elaboration and function may impose significant stress on the IM. In this study, we describe a novel role for the Cpx envelope stress response in the monitoring and regulation of the expression of NDH-I and cytochrome bo3 in EPEC. We provide evidence that the Cpx response is sensitive to the basal-level production of these protein complexes and that the inability to repress their expression in the presence of stress is lethal.
In agreement with previous microarray data, we found that the Cpx stress response regulates the expression of at least two large cytoplasmic membrane complexes in EPEC, NDH-I and cytochrome bo3 (Fig. 1) (5). The repression of nuo and cyo transcription by the Cpx response is likely mediated through direct binding of CpxR within the promoter region of these operons. The putative CpxR binding site in the cyo promoter overlaps the predicted −35 box. Therefore, CpxR likely blocks RNA polymerase binding to the cyo promoter through steric hindrance (38). However, the mechanism by which CpxR directly prevents transcription at the nuo promoter is less clear. We identified a putative CpxR binding site approximately 104 bp upstream of the nuoA transcription start site that is required for the repression of nuo-lux expression upon activation of the Cpx response (Fig. 1B and C). This putative binding site is located upstream from the predicted −35 and −10 promoter elements, suggesting that CpxR does not repress nuo transcription through steric hindrance of RNA polymerase. Upon further investigation, we identified a second putative CpxR binding site approximately 48 bp downstream of the nuoA transcription start site (Fig. 1B). Thus, it is possible that interaction between CpxR bound separately at the proximal and distal binding sites prevents nuo transcription initiation through a looping mechanism (38). Alternatively, CpxR may work with additional regulatory pathways to repress the transcription of nuo. Several lines of evidence point to a role for the ArcAB two-component system in the control of nuo transcription during envelope stress. Like CpxR, the response regulator ArcA directly represses the transcription of nuo (39, 40). However, the putative ArcA binding site within this promoter has not been reported. Inspection of the EPEC nuo promoter region with Virtual Footprint identified a putative ArcA binding site approximately 81 bp upstream of the nuoA transcription start site (Fig. 1B). As the putative ArcA and CpxR binding sites are close to one another, it is possible that these regulators work in synergy to repress the transcription of the nuo operon. Furthermore, ArcA is required for outer membrane integrity, suggesting that the ArcAB two-component system may be active during periods of envelope stress (41).
As NDH-I and cytochrome bo3 oxidase facilitate aerobic respiration, we examined oxygen consumption in different Cpx backgrounds to validate our findings. As expected, we found that constitutive activation of the Cpx response reduces oxygen consumption (Fig. 2). Surprisingly, we found that oxygen consumption was also reduced in EPEC lacking the Cpx response (Fig. 2), despite very little change in the transcription of the nuo or cyo gene cluster (Fig. 1). Under the conditions used here (mid-log-phase cells respiring in terrific broth), the respiratory defect of the ΔcpxRA mutant appears to be largely due to effects on cytochrome bo3 oxidase (Fig. S1). Taking into account the demonstrated role of the Cpx response in protein folding and degradation and the fact that proper folding of respiratory subunits is required for their assembly into complexes (33), it is possible that in the absence of the Cpx response, respiratory complexes cannot be assembled properly. As little is known about the quality control of membrane-bound respiratory complexes in either mitochondria or E. coli, potential Cpx-regulated factors involved in this process remain mysterious. While a small number of assembly factors involved in the biogenesis of iron-sulfur clusters in NDH-I have been identified, none of these are known to be regulated by the Cpx response (5, 42). Alternatively, or in addition, since the cpxRA mutant is initially able to consume oxygen at early time points in our assay (Fig. 2 and S1), perhaps the Cpx-regulated factor(s) contributes to the stability, recycling, or maintenance of these protein complexes. At present, we cannot distinguish among these possibilities.
The Cpx response plays a role in the biogenesis of other macromolecular envelope complexes, including the type IV bundle-forming pilus (13, 43). cpxR mutants display decreased activity of the bundle-forming pilus despite no change in the transcription of the bfp gene cluster. This was attributed to the decreased expression of several Cpx-regulated protein folding factors that are required for proper folding of the pilus components. Both the elaboration and retraction of type IV pili are thought to involve the extension or retraction of pilus fibers through a platform in the IM where individual pilin subunits are removed from, or added to, large pools that accumulate in the IM (44). Taken together with our hypothesis that the Cpx response may play a role in the biogenesis of membrane-bound respiratory complexes, these studies suggest that the Cpx response may play a general role in the biogenesis and/or quality control of abundant IM protein complexes.
Deletion of nuo, cyo, or both increases the resistance of the cpxR mutant to several Cpx-specific stresses (Fig. 3). One possible explanation for this result is that changes in the proton motive force (PMF) as a result of deletion of these complexes reduce the presence of stressors at the IM. NlpE is secreted into the envelope through the PMF-dependent Sec translocon, and aminoglycoside antibiotics require the PMF for uptake (11, 45). Furthermore, the expression of both nuo and cyo is decreased at alkaline pH to maintain the cytoplasmic pH (46). However, a previous report has shown that the PMF is not substantially altered in E. coli lacking both NDH-I and cytochrome bo3 (47). Therefore, changes in PMF are not likely to account for the observed resistance of these mutants to envelope stress. We propose, instead, that defects in the assembly of, or irreparable damage to, these complexes in the cpxR mutant may increase sensitivity to envelope stressors by disrupting IM integrity. In the presence of envelope stress, EPEC with a functional Cpx response represses the de novo synthesis of NDH-I and cytochrome bo3 (Fig. 1), thus reducing protein traffic within the IM. Additional Cpx-regulated factors may assist in the biogenesis or repair of existing complexes (Fig. 2) to reduce membrane damage. However, in the cpxR mutant, the expression of NDH-I and cytochrome bo3 is unchanged during stress (Fig. 1). Therefore, newly synthesized respiratory complexes may be inserted into an already damaged membrane. In the absence of Cpx-regulated assembly factors, unassembled or misassembled respiratory subunits may further disrupt IM integrity (Fig. 3). Therefore, deletion of nuo or cyo would reduce stress on the IM in the cpxR mutant. By whatever mechanism NDH-I and cytochrome bo3 exert their toxicity, these results suggest that regulation of these complexes by the Cpx response is centrally involved in adaptation to envelope stress.
Expression of NDH-I and cytochrome bo3 contributes to the basal activity of the Cpx stress response in EPEC, as indicated by the decrease in cpxP expression in bacteria lacking the nuo or cyoA operon (Fig. 4A). The mechanism by which the Cpx response might detect the presence of these protein complexes remains mysterious. One possibility is that the Cpx response detects malfunctions in these complexes that lead to the production of a Cpx-activating signal. In this regard, it is known that electron flow leads to the generation of damaging reactive oxygen species (48), which could theoretically result in damage to respiratory complexes or other protein assemblies and a Cpx-inducing signal. In agreement with this model, Bina and coworkers recently showed in Vibrio cholerae that the inability to efflux the siderophore vibriobactin, as well as the oxidative-stress-inducing agent paraquat, resulted in Cpx pathway induction in an oxygen-dependent manner (49). Additionally, Chao and Vogel (50) demonstrated that the Cpx response is activated by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) in Salmonella, suggesting that disruption of the proton-pumping activity associated with these complexes could also potentially be the source of a Cpx-activating signal.
While we cannot definitively say whether the enzymatic activities and/or the assembly of these ETC complexes is responsible for inducing the Cpx response, we believe our results support the conclusion that the Cpx envelope stress response is responsive to some aspect of their biogenesis. We found that, as reported previously by Danese and Silhavy (16), the E. coli Cpx response is not induced by CCCP (Fig. S4A and B). Further, we observed that the ability of the nuo and cyo deletions to suppress the sensitivity of a cpxR mutant to overexpression of NlpE, as well as amikacin, is additive (Fig. 3A and S2). Similarly, the impact of these mutations on Cpx pathway activity is also additive (Fig. 4A). Finally, overexpression of individual subunits of the cytochrome bo3 oxidase complex induces the Cpx response (Fig. 4B), and we found this induction to occur even in a mutant lacking the cyo operon (data not shown). Cumulatively, our findings support the hypothesis that the Cpx response detects some signal related specifically to the assembly of IM protein complexes, much as the σE envelope stress response responds to a specific signature element present in outer membrane proteins (OMPs) (51).
This model is further bolstered by the fact that the Cpx response downregulates the expression of other complexes that involve IM protein assemblies that are not involved in respiration, including genes encoding pili, flagella, and the type three secretion system (43, 52–55). During the normal biogenesis of complexes such as NDH-I and cytochrome bo3, it is possible that some subunits are not assembled correctly. Such subunits may engage in nonproductive interactions that result in activation of the Cpx response. Furthermore, depletion of the IM insertase/assembly factor YidC, which is required for the assembly of NDH-I, cytochrome bo3, and many other IM proteins, also activates the Cpx response (14, 56, 57). Finally, the expression of IM-localized proteolytic factors responsible for quality control at the IM, including HtpX and YccA, is under the control of the Cpx response (4, 15). These results suggest that the Cpx response monitors the biogenesis of membrane-bound protein complexes like NDH-I and cytochrome bo3 through a signal that is specific to IM proteins. In this regard, it is of interest that Chao and Vogel have shown that the small RNA (sRNA) CpxQ, encoded in the 3′ end of the cpxP mRNA, serves to downregulate the expression of several integral IM proteins (50). Additionally, Grabowicz et al. recently showed that downregulation of the periplasmic chaperone Skp by the CpxQ sRNA is likely needed to stem the aberrant insertion of OMPs into the cytoplasmic membrane (58). Cumulatively, these results support the conclusion that the Cpx response serves as a sentinel of IM protein biogenesis.
We also observed that the presence of these complexes contributes to the activation of the Cpx response upon entry into stationary phase (Fig. 4A). Wolfe and colleagues have shown that stationary-phase activation of the Cpx response occurs by two separate processes (59). First, consumption of the amino acids present in complex medium throughout growth increases the pH of the surrounding environment. Second, catabolism through the Pta-AckA acetogenesis pathway prevents the accumulation of an unidentified inhibitory metabolite. Either of these processes may be affected by loss of NDH-I or cytochrome bo3. nuo mutants have a growth defect in stationary phase that is in part due to their inability to catabolize multiple amino acids (60). Furthermore, inhibition of electron transport through NDH-I or cytochrome bo3 may alter metabolism in such a way that the unidentified inhibitory metabolite is produced. Whatever happens, it is clear that activation of the Cpx response upon entry into stationary phase is associated with both NDH-I and cytochrome bo3.
Overall, we have demonstrated a role for the Cpx response in the control of two large, abundant IM protein complexes (Fig. 5). It is well established that the σE envelope stress response serves as a sentinel for damage to prevalent β-barrel OMPs (2), and our work suggests that the Cpx response may function in an analogous fashion at the IM, responding to stresses that impair abundant protein complexes and threaten cellular integrity. Previous studies have linked the Cpx response to diverse processes, including peptidoglycan metabolism, biogenesis of virulence factors, motility, solute transport, protein export, and extrusion of waste (13, 43, 53, 54, 61–68). We believe that the pleiotropic phenotypes displayed by Cpx mutants may reflect widespread changes in the biogenesis of IM proteins, which may include altered energetics due to changes in NDH-I and cytochrome bo3 oxidase expression.
FIG 5.
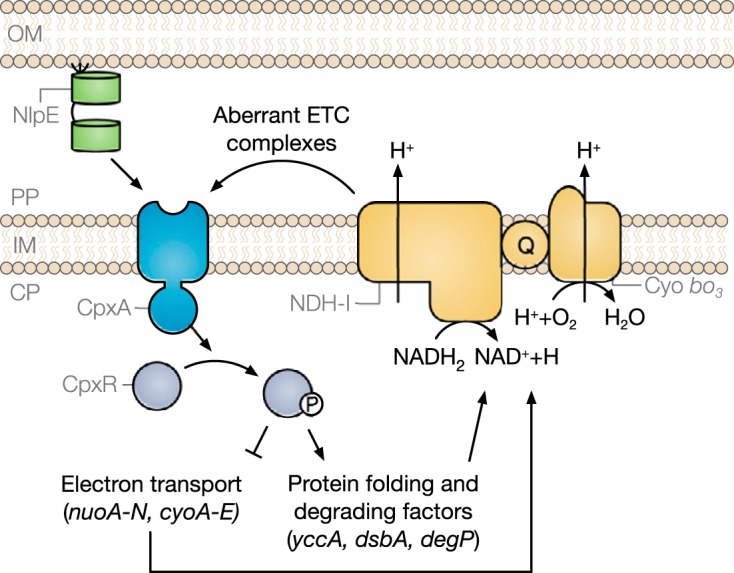
Proposed association between the Cpx envelope stress response and the ETC. Upon activation of the Cpx response, CpxR represses the transcription of the operons encoding the NDH-I and cytochrome bo3 respiratory complexes. The Cpx response may further regulate the biogenesis of these complexes at the posttranscriptional level through increased expression of protein-folding and -degrading factors. These complexes also contribute to Cpx pathway activity. OM, outer membrane; PP, periplasm; CP, cytoplasm; Q, quinone; Cyo, cytochrome.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacteria were routinely cultured in LB-Lennox broth at 37°C with aeration at 225 rpm. Isopropyl-β-d-thiogalactopyranoside (IPTG; Invitrogen) was added to a concentration of 0.1 or 1 mM, as indicated. Unless otherwise stated, antibiotics (Sigma) were used as required at the following concentrations: amikacin, 3 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml.
Strain and plasmid construction.
All EPEC mutants were constructed by allelic exchange (69). Regions of DNA approximately 1 kb upstream and downstream of the target site were amplified by PCR and joined by overlap extension PCR (primer sequences, including restriction sites, are listed in Table S2) (70). The full-length PCR products were digested with the XbaI, KpnI, or SacI (Invitrogen) restriction endonuclease and ligated into pRE112. Constructs were transferred onto the EPEC chromosome as previously described (71). Mutations were confirmed by PCR or DNA sequencing.
Luminescent reporters of NDH-I and cytochrome bo3 transcription were constructed as previously described (72). Briefly, the promoter region of each operon was amplified from the E2348/69 genome with the primers listed in Table S2. Gel-purified products were digested with EcoRI, BamHI, or PvuI (Invitrogen) and ligated upstream of the luxABCDE operon in the pJW15 plasmid. The predicted CpxR binding site in the pJW15-Pnuo reporter was mutated by overlap extension PCR (70). The mutated promoter DNA was digested and ligated into pJW15 as described above. PCR and DNA sequencing verified the correct insertion of the promoter sequences.
Luminescence assay.
Bacteria were grown overnight in LB-Lennox broth at 37°C with aeration and then subcultured 1:100 into 2 ml of fresh LB-Lennox broth in a 13- by 100-mm glass test tube at 37°C with aeration. To measure the transcription of NDH-I and cytochrome bo3, 0.1 mM IPTG was added at the time of subculture to induce NlpE expression from the pCA-nlpE plasmid. Bacteria were grown to an optical density at 600 nm (OD600) of 0.4 to 0.5, at which point luminescence was measured. To determine Cpx pathway activity in EPEC, expression of cpxP from pJW25 (Table S1) was measured. Bacteria were grown overnight as described above, subcultured at a dilution factor of 1:100 into 10 ml of LB-Lennox broth in a 125-ml Erlenmeyer flask, and then grown at 37°C with aeration. Luminescence and OD600 were measured every 2 h for 8 h postsubculture as previously described (54). Luminescence values were standardized to the OD600 of the same culture to account for differences in cell numbers between samples. All luminescence assays were repeated at least twice in quintuplicate (nuo-lux and cyo-lux) or triplicate (cpxP-lux).
Oxygen consumption.
Bacteria were grown overnight in 5 ml of LB-Lennox broth at 37°C (strains E2348/69 and RG222) or 30°C (strain ALN195) with shaking at 225 rpm. Overnight cultures were diluted by a factor of 1:100 into 10 ml of terrific broth without antibiotics in a 125-ml Erlenmeyer flask and grown to an OD600 of 0.35 at 37°C with shaking at 200 rpm. Cells were washed twice with phosphate-buffered saline (Sigma) and suspended in 1 ml of phosphate-buffered saline at a density of ∼4 × 107 CFU/ml in a closed 1-ml microrespiration chamber (Unisense). After the baseline oxygen concentration was established, respiration was initiated by the addition of 1% terrific broth. The oxygen concentration was measured every 30 s for 45 min with an oxygen MicroOptode sensor (Unisense). The oxygen concentration at each time point was standardized to the oxygen concentration immediately before the addition of terrific broth. A magnetic stirrer was used during the assay to ensure that oxygen was distributed throughout the microrespiration chamber. The data shown are representative of three replicate experiments.
Sensitivity assays.
To determine sensitivity to nlpE overexpression, bacteria containing nlpE overexpression vector pCA-nlpE or control vector pCA-24N were grown overnight in 5 ml of LB-Lennox broth at 37°C with aeration and subcultured 1:100 into 5 ml of fresh LB-Lennox broth at 37°C with aeration. IPTG at 1 mM was added at the early exponential phase to induce the expression of NlpE from pCA-nlpE, and cultures were grown for an additional 4.5 h. CFU counts were measured by serial dilution and growth on LB-Lennox agar. The number of CFU/ml was calculated by standardizing the number of resulting colonies to the dilution factor. NlpE sensitivity assays were performed twice in triplicate. To determine sensitivity to aminoglycoside antibiotics and alkaline pH, overnight cultures were standardized to an OD600 of 1, serially diluted, and plated on plain LB-Lennox agar, agar containing 1.5 μg/ml amikacin, agar buffered to pH 7.0, or agar buffered to pH 9.5. Agar was buffered to pH 7.0 or 9.5 with sodium hydroxide. Plates were incubated overnight at 37°C. The amikacin and pH sensitivity assays shown represent at least two replicate experiments.
Genetic screen.
To identify genes involved in modulation of the activity of the Cpx response in E. coli, 176 Cpx-regulated envelope-localized proteins were screened on the basis of color variation on lactose MacConkey agar with a cpxP-lacZ reporter. From a previously published microarray study that characterizing the Cpx regulon upon NlpE overexpression in E. coli, genes whose expression was regulated at least 2-fold were identified (5). The candidate pool was further narrowed down to genes encoding envelope-localized proteins by referring to their cellular localization listed in the Ecocyc database (73). The genetic screen was designed and performed in accordance with a previously described methodology (72). For each candidate tested, its overexpression plasmid from the ASKA library was extracted and transformed into TR50 (Table S1), which carries a chromosomal cpxP-lacZ reporter. Four single colonies of the resulting transformants, along with the control TR50 (pCA-24N), were patched onto lactose MacConkey plates supplemented with 0.1 mM IPTG to induce expression from the plasmid. In comparison to TR50 (pCA-24N), brighter red colonies indicated high levels of lacZ transcription and pink or white colonies indicated low levels of lacZ expression. The observed inhibitory or activating phenotype of candidates that showed changed Cpx activity was further confirmed by β-galactosidase assay to quantify the activity of the Cpx pathway.
β-Galactosidase assays.
β-Galactosidase activity was measured in microtiter plates as previously described (74). Bacteria were grown overnight in LB-Lennox broth at 37°C (wild-type TR50 strains) or 30°C (cpxA mutant TR50 strains) with aeration at 225 rpm. Overnight cultures then were subcultured in a 1:100 dilution into 2 ml of fresh LB-Lennox broth in a 13- by 100-mm glass test tube with aeration. To induce protein expression from pCA-based plasmids, 0.1 mM IPTG was added 1 h after subculture. Bacteria were collected by centrifugation when the OD600 reached 0.4 to 0.6 and resuspended in 2 ml of freshly prepared buffer Z (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, and 1 mM MgSO4 · 7H2O containing 270 μl of β-mercaptoethanol). A 250-μl volume of cell mixture was then transferred to a 96-well microtiter plate, and the OD600 was read with a plate reader (PerkinElmer). The remaining cells were lysed for 10 min with 2 drops of chloroform and 1 drop of 0.1% SDS, and the cellular debris was removed by centrifugation. A 50-μl volume of 10 mg/ml o-nitrophenyl-β-d-galactopyranoside (Sigma) was then added to diluted cell lysate with a 5-μl aliquot of lysed cell mixture and 195 μl of buffer Z in a 96-well plate to initiate the reaction. The A420 was read 20 times over approximately 30 min in the plate reader, and Miller units were calculated (75). Experiments were done in triplicate three times.
Statistical analysis.
Statistical analysis was performed with Prism version 7.0c (GraphPad Software). The activities of transcriptional reporters were compared by one-way analysis of variance, followed by Sidak's multiple-comparison test.
Supplementary Material
ACKNOWLEDGMENTS
This work, including the efforts of Tracy L. Raivio, Randi L. Guest, and Julia L. Wong, was funded by operating grant MOP 342982 from the Canadian Institutes of Health Research. The work of Tracy L. Raivio and Junshu Wang was funded by discovery grant RGPIN 238422-2013 from the Natural Sciences and Engineering Research Council.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00153-17.
For a commentary on this article, see https://doi.org/10.1128/JB.00433-17.
REFERENCES
- 1.Weiner JH, Li L. 2008. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim Biophys Acta 1778:1698–1713. doi: 10.1016/j.bbamem.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 2.MacRitchie DM, Raivio TL. 2009. Envelope stress responses. EcoSal Plus 2013 doi: 10.1128/ecosalplus.5.4.7. [DOI] [PubMed] [Google Scholar]
- 3.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol 191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raivio TL, Leblanc SKD, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangaiah D, Zhang X, Fortney KR, Baker B, Liu Y, Munson RS, Spinola SM. 2013. Activation of CpxRA in Haemophilus ducreyi primarily inhibits the expression of its targets, including major virulence determinants. J Bacteriol 195:3486–3502. doi: 10.1128/JB.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acosta N, Pukatzki S, Raivio TL. 2015. The Vibrio cholerae Cpx envelope stress response senses and mediates adaptation to low iron. J Bacteriol 197:262–276. doi: 10.1128/JB.01957-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun S, Lee E-G, Kim S-Y, Shin JM, Jung WS, Oh D-B, Lee SY, Kwon O. 2015. The CpxRA two-component system is involved in the maintenance of the integrity of the cell envelope in the rumen bacterium Mannheimia succiniciproducens. Curr Microbiol 70:103–109. doi: 10.1007/s00284-014-0686-5. [DOI] [PubMed] [Google Scholar]
- 9.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol 181:5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tschauner K, Hörnschemeyer P, Müller VS, Hunke S. 2014. Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS One 9:e107383. doi: 10.1371/journal.pone.0107383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J 16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nevesinjac AZ, Raivio TL. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J Bacteriol 187:672–686. doi: 10.1128/JB.187.2.672-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Kuhn A, Dalbey RE. 2010. Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J Bacteriol 192:2193–2209. doi: 10.1128/JB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662. doi: 10.1046/j.1365-2443.2002.00554.x. [DOI] [PubMed] [Google Scholar]
- 16.Danese PN, Silhavy TJ. 1998. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 19.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 20.Raivio TL, Laird MW, Joly JC, Silhavy TJ. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the Cpx envelope stress response. Mol Microbiol 37:1186–1197. doi: 10.1046/j.1365-2958.2000.02074.x. [DOI] [PubMed] [Google Scholar]
- 21.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 22.Price CE, Driessen AJ. 2010. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim Biophys Acta 1803:748–766. doi: 10.1016/j.bbamcr.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Leif H, Sled VD, Ohnishi T, Weiss H, Friedrich T. 1995. Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur J Biochem 230:538–548. [DOI] [PubMed] [Google Scholar]
- 24.Hofhaus G, Weiss H, Leonard K. 1991. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (complex I). J Mol Biol 221:1027–1043. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 25.Guénebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. 1998. Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I). J Mol Biol 276:105–112. doi: 10.1006/jmbi.1997.1518. [DOI] [PubMed] [Google Scholar]
- 26.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. 2013. Crystal structure of the entire respiratory complex I. Nature 494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. 1993. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli: organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol 233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 28.Leif H, Weidner U, Berger A, Spehr V, Braun M, van Heek P, Friedrich T, Ohnishi T, Weiss H. 1993. Escherichia coli NADH dehydrogenase I, a minimal form of the mitochondrial complex I. Biochem Soc Trans 21:998–1001. doi: 10.1042/bst0210998. [DOI] [PubMed] [Google Scholar]
- 29.Ghaim JB, Tsatsos PH, Katsonouri A, Mitchell DM, Salcedo-Hernandez R, Gennis RB. 1997. Matrix-assisted laser desorption ionization mass spectrometry of membrane proteins: demonstration of a simple method to determine subunit molecular weights of hydrophobic subunits. Biochim Biophys Acta 1330:113–120. doi: 10.1016/S0005-2736(97)00127-2. [DOI] [PubMed] [Google Scholar]
- 30.Hosler JP, Ferguson-Miller S, Calhoun MW, Thomas JW, Hill J, Lemieux L, Ma J, Georgiou C, Fetter J, Shapleigh J, Tecklenburg MMJ, Babcock GT, Gennis RB. 1993. Insight into the active-site structure and function of cytochrome oxidase by analysis of site-directed mutants of bacterial cytochrome aa3 and cytochrome bo. J Bioenerg Biomembr 25:121–136. doi: 10.1007/BF00762854. [DOI] [PubMed] [Google Scholar]
- 31.Musatov A, Ortega-Lopez J, Demeler B, Osborne JP, Gennis RB, Robinson NC. 1999. Detergent-solubilized Escherichia coli cytochrome bo3 ubiquinol oxidase: a monomeric, not a dimeric complex. FEBS Lett 457:153–156. doi: 10.1016/S0014-5793(99)01020-0. [DOI] [PubMed] [Google Scholar]
- 32.Chepuri V, Lemieux L, Au DC, Gennis RB. 1990. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem 265:11185–11192. [PubMed] [Google Scholar]
- 33.Stenberg F, von Heijne G, Daley DO. 2007. Assembly of the cytochrome bo3 complex. J Mol Biol 371:765–773. doi: 10.1016/j.jmb.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 34.García-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J Bacteriol 176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 36.DiGiuseppe PA, Silhavy TJ. 2003. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol 185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. 2016. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 39.Bongaerts J, Zoske S, Weidner U, Unden G. 1995. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol 16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 40.Salmon KA, Hung S-P, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. 2005. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem 280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 41.Wan F, Mao Y, Dong Y, Ju L, Wu G, Gao H. 2015. Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis. Sci Rep 5:10228. doi: 10.1038/srep10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedrich T, Dekovic DK, Burschel S. 2016. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (respiratory complex I). Biochim Biophys Acta 1857:214–223. doi: 10.1016/j.bbabio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. 2010. The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus. Mol Microbiol 76:1095–1110. doi: 10.1111/j.1365-2958.2010.07145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leighton TL, Buensuceso RNC, Howell PL, Burrows LL. 2015. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17:4148–4163. doi: 10.1111/1462-2920.12849. [DOI] [PubMed] [Google Scholar]
- 45.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer LM, Yohannes E, BonDurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engl C, Beek AT, Bekker M, de Mattos JT, Jovanovic G, Buck M. 2011. Dissipation of proton motive force is not sufficient to induce the phage shock protein response in Escherichia coli. Curr Microbiol 62:1374–1385. doi: 10.1007/s00284-011-9869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imlay JA, Fridovich I. 1991. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem 266:6957–6965. [PubMed] [Google Scholar]
- 49.Kunkle DE, Bina XR, Bina JE. 2017. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8:e00126-17. doi: 10.1128/mBio.00126-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chao Y, Vogel J. 2016. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–363. doi: 10.1016/j.molcel.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 51.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61–71. doi: 10.1016/S0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 52.Hernday AD, Braaten BA, Broitman-Maduro G, Engelberts P, Low DA. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol Cell 16:537–547. doi: 10.1016/j.molcel.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 53.De Wulf P, Kwon O, Lin EC. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol 181:6772–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacRitchie DM, Ward JD, Nevesinjac AZ, Raivio TL. 2008. Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect Immun 76:1465–1475. doi: 10.1128/IAI.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacRitchie DM, Buelow DR, Price NL, Raivio TL. 2008. Two-component signaling and Gram negative envelope stress response systems. Adv Exp Med Biol 631:80–110. doi: 10.1007/978-0-387-78885-2_6. [DOI] [PubMed] [Google Scholar]
- 56.van der Laan M, Urbanus ML, Hagen-Jongman Ten CM, Nouwen N, Oudega B, Harms N, Driessen AJM, Luirink J. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc Natl Acad Sci U S A 100:5801–5806. doi: 10.1073/pnas.0636761100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price CE, Driessen AJM. 2010. Conserved negative charges in the transmembrane segments of subunit K of the NADH:ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J Biol Chem 285:3575–3581. doi: 10.1074/jbc.M109.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabowicz M, Koren D, Silhavy TJ. 2016. The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. mBio 7:e00312-16. doi: 10.1128/mBio.00312-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfe AJ, Parikh N, Lima BP, Zemaitaitis B. 2008. Signal integration by the two-component signal transduction response regulator CpxR. J Bacteriol 190:2314–2322. doi: 10.1128/JB.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prüss BM, Nelms JM, Park C, Wolfe AJ. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol 176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernal-Cabas M, Ayala JA, Raivio TL. 2015. The Cpx envelope stress response modifies peptidoglycan cross-linking via the l,d-transpeptidase LdtD and the novel protein YgaU. J Bacteriol 197:603–614. doi: 10.1128/JB.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delhaye A, Collet J-F, Laloux G. 2016. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7:e00047-16. doi: 10.1128/mBio.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacRitchie DM, Acosta N, Raivio TL. 2012. DegP is involved in Cpx-mediated posttranscriptional regulation of the type III secretion apparatus in enteropathogenic Escherichia coli. Infect Immun 80:1766–1772. doi: 10.1128/IAI.05679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plate CA. 1976. Mutant of Escherichia coli defective in response to colicin K and in active transport. J Bacteriol 125:467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plate CA, Seely SA, Laffler TG. 1986. Evidence for a protonmotive force related regulatory system in Escherichia coli and its effects on lactose transport. Biochemistry 25:6127–6132. doi: 10.1021/bi00368a044. [DOI] [PubMed] [Google Scholar]
- 66.Plate CA, Suit JL. 1981. The eup genetic locus of Escherichia coli and its role in H+/solute symport. J Biol Chem 256:12974–12980. [PubMed] [Google Scholar]
- 67.Taylor DL, Bina XR, Slamti L, Waldor MK, Bina JE. 2014. Reciprocal regulation of resistance-nodulation-division efflux systems and the Cpx two-component system in Vibrio cholerae. Infect Immun 82:2980–2991. doi: 10.1128/IAI.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishino K, Yamasaki S, Hayashi-Nishino M, Yamaguchi A. 2010. Effect of NlpE overproduction on multidrug resistance in Escherichia coli. Antimicrob Agents Chemother 54:2239–2243. doi: 10.1128/AAC.01677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 70.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 71.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59:4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong JL, Vogt SL, Raivio TL. 2013. Using reporter genes and the Escherichia coli ASKA overexpression library in screens for regulators of the Gram negative envelope stress response. Methods Mol Biol 966:337–357. doi: 10.1007/978-1-62703-245-2_21. [DOI] [PubMed] [Google Scholar]
- 73.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buelow DR, Raivio TL. 2005. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J Bacteriol 187:6622–6630. doi: 10.1128/JB.187.19.6622-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem 270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.