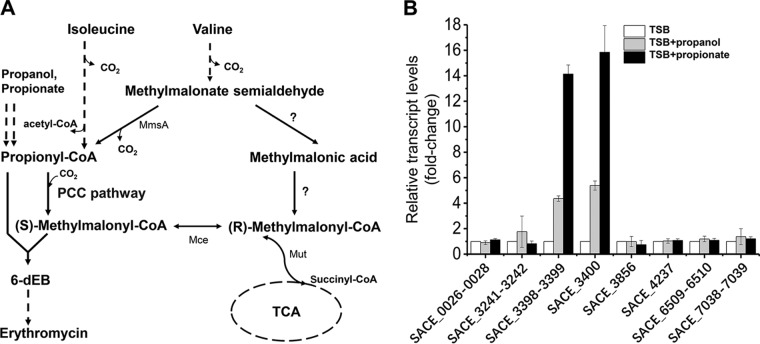
FIG 1.
pccBC (SACE_3398–3399) and pccA (SACE_3400) operons are induced by propanol and propionate in S. erythraea. (A) In S. erythraea, branched-chain amino acids (BCAA) are a source of propionyl-CoA and methylmalonyl-CoA, key precursors in erythromycin biosynthesis. Valine and isoleucine were metabolized to propionyl-CoA through the degradation pathways. Propanol and propionate are two important external sources of propionyl-CoA, especially in the production of erythromycin. The metabolism of propionyl-CoA to (2S)-methylmalonyl–CoA is via the PCC pathway in S. erythraea, which plays an important role in propionyl-CoA assimilation when n-propanol or valine is added for high production of erythromycin. Enzymes that transform methylmalonate-semialdehyde to (R)-methylmalonyl–CoA (indicated by question marks) have not yet been investigated in S. erythraea. (B) Total RNA of S. erythraea NRRL2338 was extracted after 36 h of growth (see Fig. S3 in the supplemental material) in TSB medium, TSB supplied with 1% (vol/vol) n-propanol, and TSB supplied with 20 mM propionate. Relative transcript levels were normalized to the 16S rRNA. The transcription value of each gene in S. erythraea in TSB medium was arbitrarily normalized to 1. Error bars represent the standard deviations from three biological replicates. MmsA, methylmalonate-semialdehyde dehydrogenase (acylating); Mut, methylmalonyl-CoA mutase; Mce, methylmalonyl-CoA epimerase.