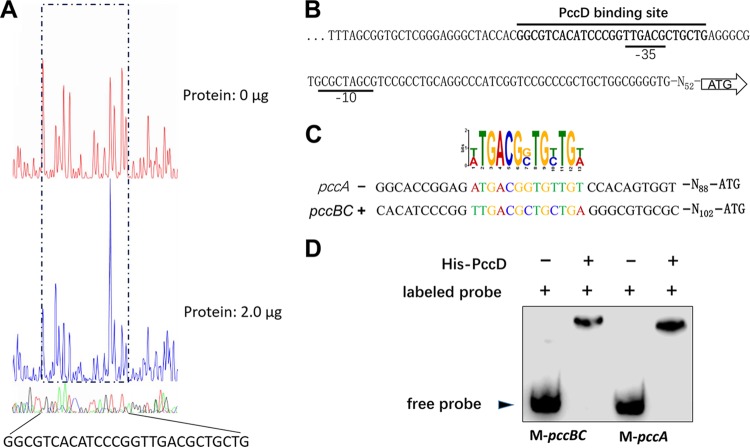
FIG 4.
Identification of a PccD-binding site in pccBC and pccA operons. (A) Electropherograms of a DNase I digest of pccBC promoter probe incubated without (top) or with (bottom) 2.0 μg of His-PccD. The nucleotide sequence protected by His-PccD is indicated below. (B) Protected sequence stretches in the upstream regions of pccBC. Black lines indicate the regions of DNase I protection. The −35 and −10 sites were predicted by the Softberry web tool. (C) Analysis of the PccD-binding motif of pccA using MEME. The standard code of the WebLogo server is shown at the top. (D) Verification of PccD-binding motif by EMSA. Synthetic probes M-pccBC and M-pccA containing PccD-binding site in promoters of pccBC and pccA operons, respectively. The DNA probe was incubated with a protein concentration of 5 μM.