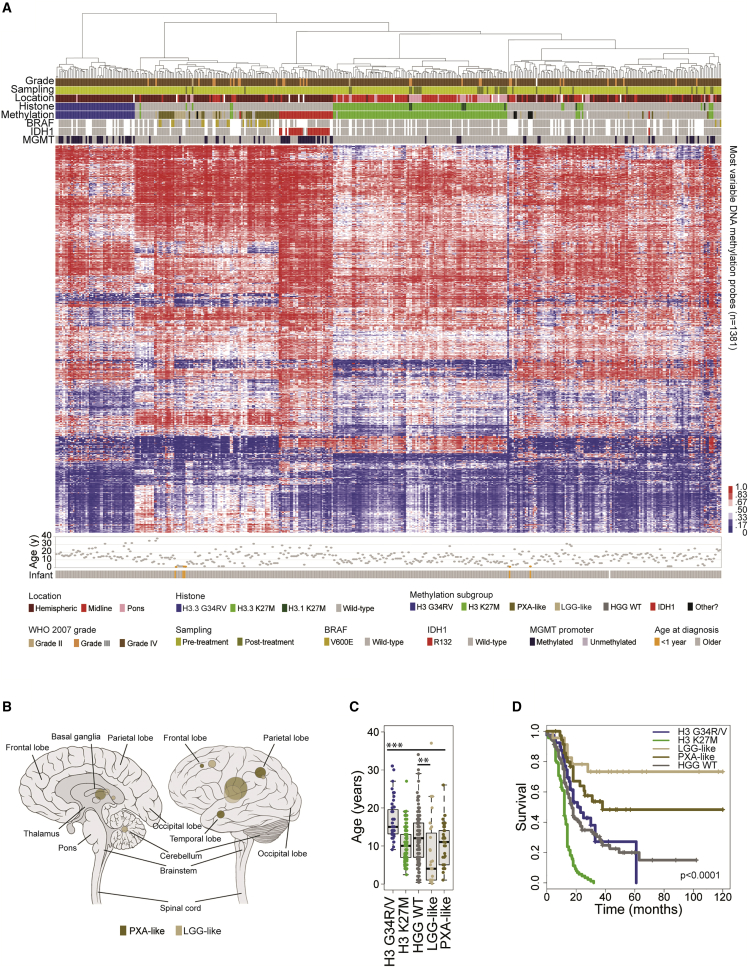
Figure 2.
Methylation-based Subclassification of pHGG/DIPG
(A) Unsupervised hierarchical clustering and heatmap representation of β values for 441 samples profiled on the Illumina 450k BeadArray platform (red, high; blue, low). Samples are arranged in columns clustered by most variable 1,381 classifier probes. Age at diagnosis is provided below. Clinicopathological and molecular annotations are provided as bars according to the included key.
(B) Anatomical location of methylation-defined PXA-like (n = 43) and LGG-like (n = 27) cases. Left, sagittal section showing internal structures; right, external view highlighting cerebral lobes. Dark gold, PXA-like; tan, LGG-like. Radius of circle is proportional to the number of cases. Lighter shaded circles represent a non-specific designation of hemispheric, midline, or brainstem.
(C) Boxplot showing age at diagnosis of included cases, separated by simplified methylation subclass (n = 440). The thick line within the box is the median, the lower and upper limits of the boxes represent the first and third quartiles, and the whiskers 1.5× the interquartile range. ∗∗∗Adjusted p < 0.0001 for all H3 G34R/V pairwise comparisons, t test; ∗∗adjusted p < 0.01 for LGG-like versus WT, t test.
(D) Kaplan-Meier plot of overall survival of cases separated by simplified methylation subclass, p value calculated by the log rank test (n = 307). See also Figure S2 and Table S2.