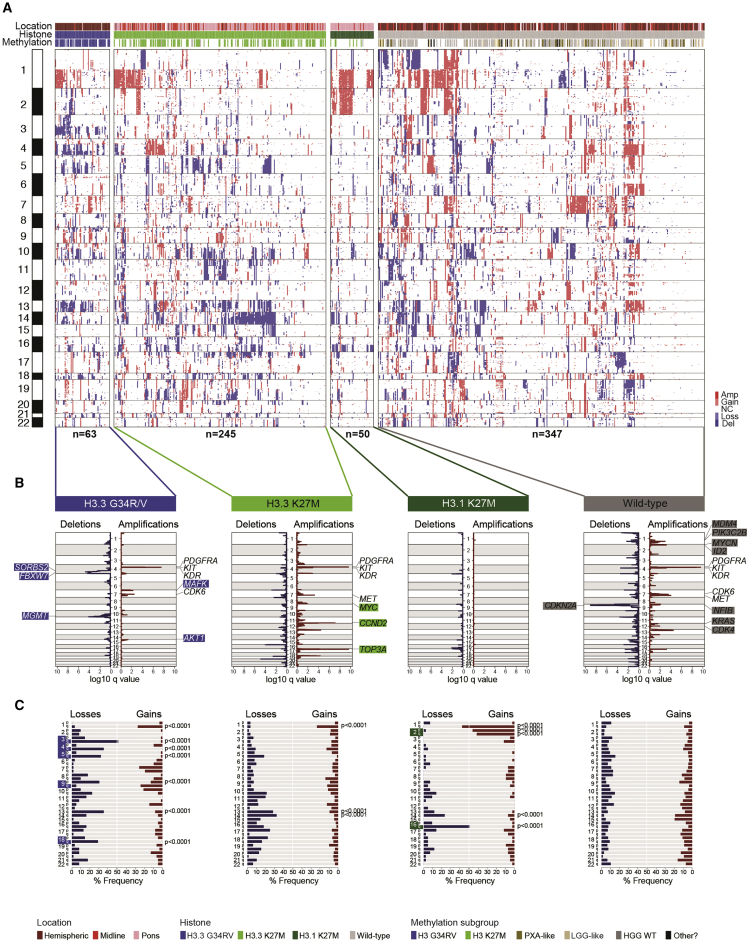
Figure 4.
Subgroup-specific Copy-Number Changes in pHGG/DIPG
(A) Heatmap representation of segmented DNA copy number for 705 pHGG/DIPG separated for known histone mutation subgroup (dark red, amplification; red, gain; dark blue, deletion; blue, loss). Samples are arranged in columns clustered by gene-level data across the whole genome. Clinicopathological and molecular annotations are provided as bars according to the included key.
(B) GISTIC analysis of focal amplifications and deletions for histone mutation subgroups. Log10 values are plotted across the genome for both amplifications (dark red) and deletions (dark blue), with significantly enriched events labeled by likely driver genes. Subgroup-specific genes are highlighted by the appropriate color.
(C) Barplot of frequency of whole chromosomal arm gains (red) and losses (blue) for each subgroup. Significantly enriched alterations (p < 0.0001, Fisher’s exact test) are labeled, with subgroup-specific arm changes highlighted by the appropriate color. See also Figure S4 and Table S4.