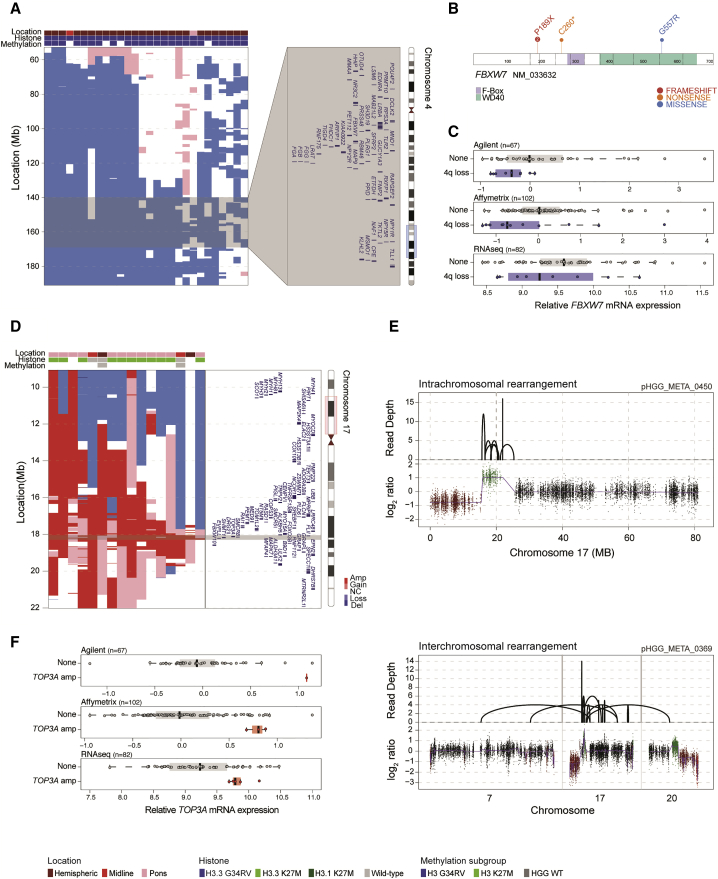
Figure 5.
Alterations Targeting FBXW7 in H3.3G34R/V pHGG and TOP3A in H3.3K27M DIPG
(A) Segmented exon-level DNA copy-number heatmaps for 4q loss in H3.3G34R/V tumors (dark red, amplification; red, gain; dark blue, deletion; blue, loss; n = 28). An ideogram of chromosome 4 is provided indicating enlarged genome browser view and genes within common regions targeted across samples (gray). Clinicopathological and molecular annotations are provided as bars according to the included key.
(B) Cartoon representation of amino acid position for four somatic mutations found in FBXW7, colored by annotated functional domains and numbers provided for recurrent variants.
(C) Boxplots representing gene expression differences between FBXW7 lost/mutated cases (blue) and those with normal copy/WT (gray) in three independent gene expression platform datasets. The thick line within the box is the median, the lower and upper limits of the boxes represent the first and third quartiles, and the whiskers 1.5× the interquartile range.
(D) Segmented exon-level DNA copy-number heatmaps for 17p11.2 amplification in predominantly H3.3K27M DIPG (dark red, amplification; red, gain; dark blue, deletion; blue, loss; n = 17). Chromosome 17 ideogram is provided indicating enlarged genome browser view and genes within common regions targeted across samples (gray). Clinicopathological and molecular annotations are provided as bars according to the included key.
(E) Sequencing coverage (top) and log2 ratio plot (bottom) for chromosomes 7, 17, and 20 for two cases, showing complex intra- or inter-chromosomal rearrangements leading to specific copy-number amplification of TOP3A.
(F) Boxplots representing gene expression differences between TOP3A amplified cases (red) and those with normal copy (gray) in three independent gene expression platform datasets. The thick line within the box is the median, the lower and upper limits of the boxes represent the first and third quartiles, and the whiskers 1.5× the interquartile range. See also Figure S5 and Table S5.