Figure 7.
Integrated Pathway Analysis of pHGG/DIPG
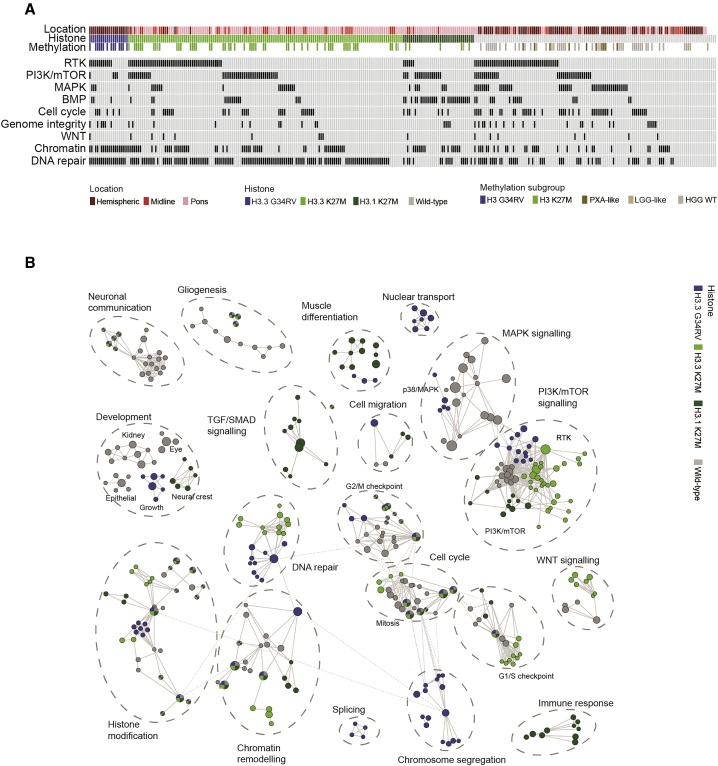
(A) Oncoprint-style representation of an integrated annotation of somatic mutations and DNA copy-number changes in one or more of nine commonly targeted pathways in 326 pHGG/DIPG (n ≥ 6, frequency barplot on the right). Samples are arranged in columns with pathways labeled along rows. Clinicopathological and molecular annotations are provided as bars according to the included key.
(B) Pathway enrichment analysis of pHGG/DIPG subgroups. Distinct pathways and biological processes between the subgroups are colored appropriately (FDR q < 0.01). Nodes represent enriched gene sets, which are grouped and annotated by their similarity according to related gene sets. Node size is proportional to the total number of genes within each gene set. The illustrated network map was simplified by manual curation to remove general and uninformative sub-networks. See also Figure S7 and Table S7.