Extended Data Figure 8.
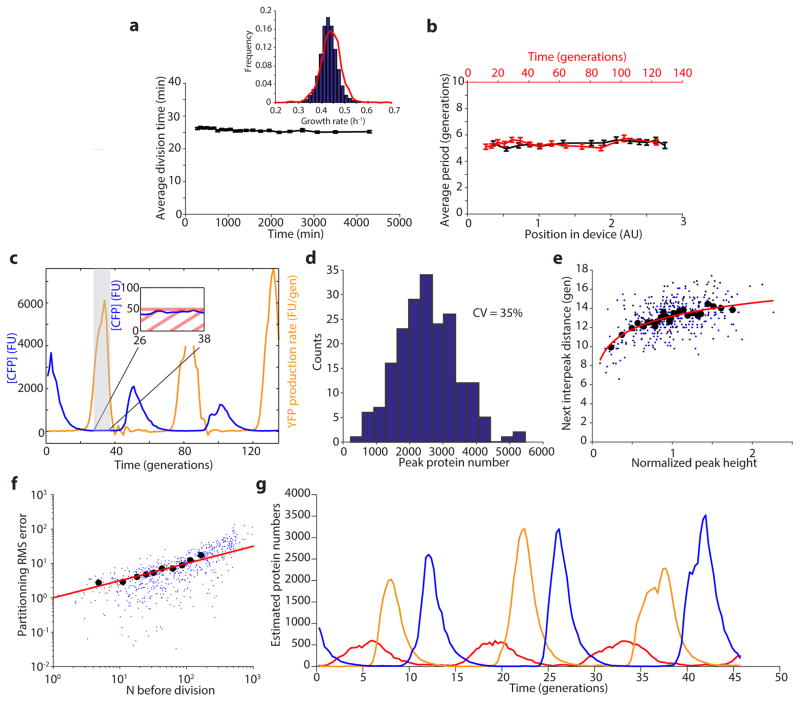
Characterization of the microfluidic device and of the oscillations. a) The average division time of the integrated repressilator (LPT25) is constant over time. The inset shows the distribution of growth rates of two independent experiments (45,828 and 9,135 points are shown in the blue and red distributions, respectively), with a slight difference in the mean (~ 1%). b) The period of the oscillations is constant in space (position in field of view, distance to inlets and outlets and different media channels) and time. Each point represents a bin of 400 (a) or 100 (b) points, with the error bars indicating standard error of the mean. c) The induction/repression switch of CI (reported by YFP) occurs when the transcriptional reporter for TetR (CFP) is below the detection limit. Typical time trace of multireporter repressilator without repressor degradation and with PLtet-peptide-asv plasmid (ΔclpXP, LPT117). The production rate of YFP is shown alongside the CFP concentration. The inset shows that the switch from induction to repression occurs below the detection limit of ~ 50 FU. d) The distribution of peak amplitude of the repressilator without degradation but with titration sponge shows significant heterogeneity (LPT64, CV of 35%). e) The peak amplitude has a small influence on the next period, due to exponential dilution. The red line shows a fit to y = 1.99 * log(x)+13.18 and explains 25% of the variance in the periods. Black circles are bins of 15 points of black dots (LPT64) and blue dots (LPT156). f) Estimating partitioning root mean squared (RMS) errors at cell division during the dilution phase showed that it scaled binomially, and allowed us to roughly estimate a fluorescence units to protein scaling factor. Black circles are bins of 50 blue dots (LPT64). The red line shows the fit (after conversion) to , where ni is the number of proteins in the daughters right after division, and N = n1 + n2 the number in the mother cell. g) Typical time trace of triple reporter repressilator without degradation with titration sponge (LPT127) in estimated protein numbers (concentration × average cell size).