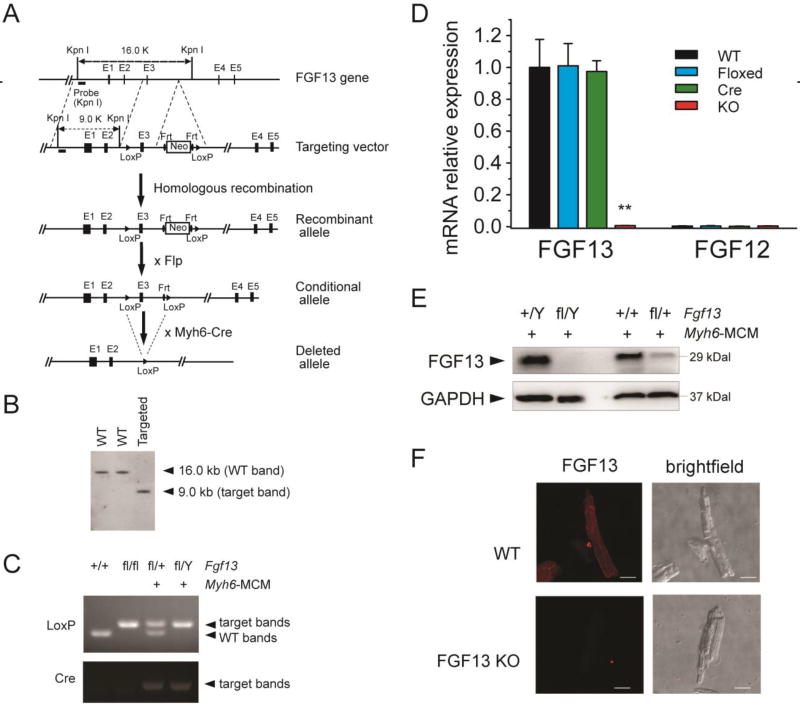
Fig. 1.
Generation of FGF13 cardiac conditional knockout mice using Cre-LoxP system. (A) Structure of the mouse Fgf13 genomic locus (line 1; black boxes represent exons); targeting vector to introduce a neomycin selection cassette and floxed exon 3 (line 2); and the recombined Fgf13 allele (line 3). Crossing with a FLP deleter yields a conditional allele (line 4). Subsequent cross with Myh6-Cre generates a tamoxifen-inducible cardiac-specific KO (line 5). (B) Southern blot of genomic DNA from targeted ES cell clone and two wild-type ES cell clones, hybridized with 5’ genomic probes. (C) PCR genotyping of wild-type (+/+), homozygous flox (fl/fl), heterozygous (fl/+; Myh6-MCM+) and hemizygous (fl/Y; Myh6-MCM+) FGF13 mice using genomic DNA prepared from tail biopsies. (D) RT-qPCR of FGF13 and FGF12B in Fgf13fl/Y; Myh6-MCM+/− (KO) compared to wild type (WT), floxed FGF13 (Floxed), and Myh6-MCM (Cre) showing ~99% reduction of FGF13 mRNA and lack of compensation by FGF12B. The RTqPCR was performed one week after tamoxifen induction. Ct values were corrected with GAPDH and normalized to FGF13 in WT. **indicates p<0.01 compared to WT. n=3. (E) Representative immunoblots of FGF13 protein expression in ventricular myocytes from Myh6-MCM, hemizygous, and heterozygous Fgf13 KO mice. (F) Confocal images of FGF13 immunocytochemistry (left panels) with corresponding bright-field images (right panels) in WT (upper panels) and Fgf13 KO (lower panels) mice. Scale bar, 25 µm.