Figure 1.
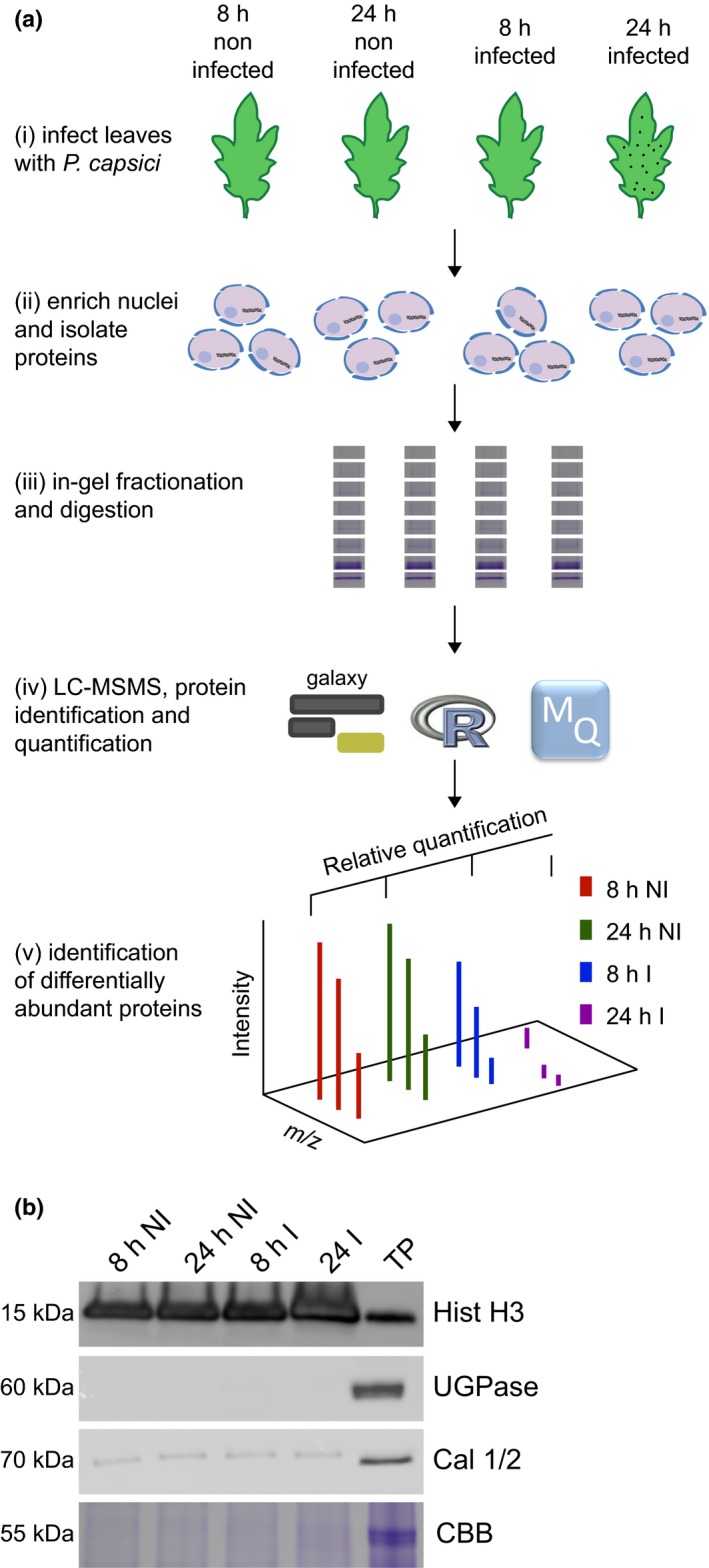
A simple workflow combining nuclear enrichment with quantitative mass spectrometry allows the study of host nuclear processes during infection. (a) Method overview. Detached leaves from 4‐wk‐old tomato plants were spray‐inoculated with Phytophthora capsici zoospores at a concentration of 500 000 spores ml−1, or water as a control (i). Leaves were harvested 8 and 24 h post‐infection and subject to nuclear enrichment (ii). Nuclear protein extracts were generated and fractionated in‐gel and digested with trypsin (iii). Peptide samples were subjected to liquid chromatography−tandem mass spectrometry (LC‐MSMS) analysis and peptide identification and label‐free quantification was carried out using the maxquant and perseus software packages, to identify proteins that were differentially expressed during infection. Nuclear predictions were performed using prediction software on our in‐house Galaxy server as described in the Methods. Subsequent data filtering was completed using the r software package (iv and v). Three independent biological replicates were generated. I, infected samples; NI, noninfected samples (v). (b) Successful enrichment of nuclear proteins demonstrated by western blotting with anti‐histone H3 antibody and subcellular markers. Protein extracts from enriched nuclear samples were compared to a total protein extract (TP) from tomato leaves. Protein concentrations were adjusted for equal loading. Nonnuclear contamination was assessed by probing with anti‐UDP‐glucose pyrophosphorylase (UGPase) antibody (cytoplasm) and calnexin homologue 1/2 antibody (endoplasmic reticulum). Samples were also run on gels and stained with Coomassie brilliant blue (CBB) to assess protein loading and Rubisco abundance. The figure shows the results from a single biological replicate. Blots for all three replicates are provided in Supporting Information Fig. S2.
