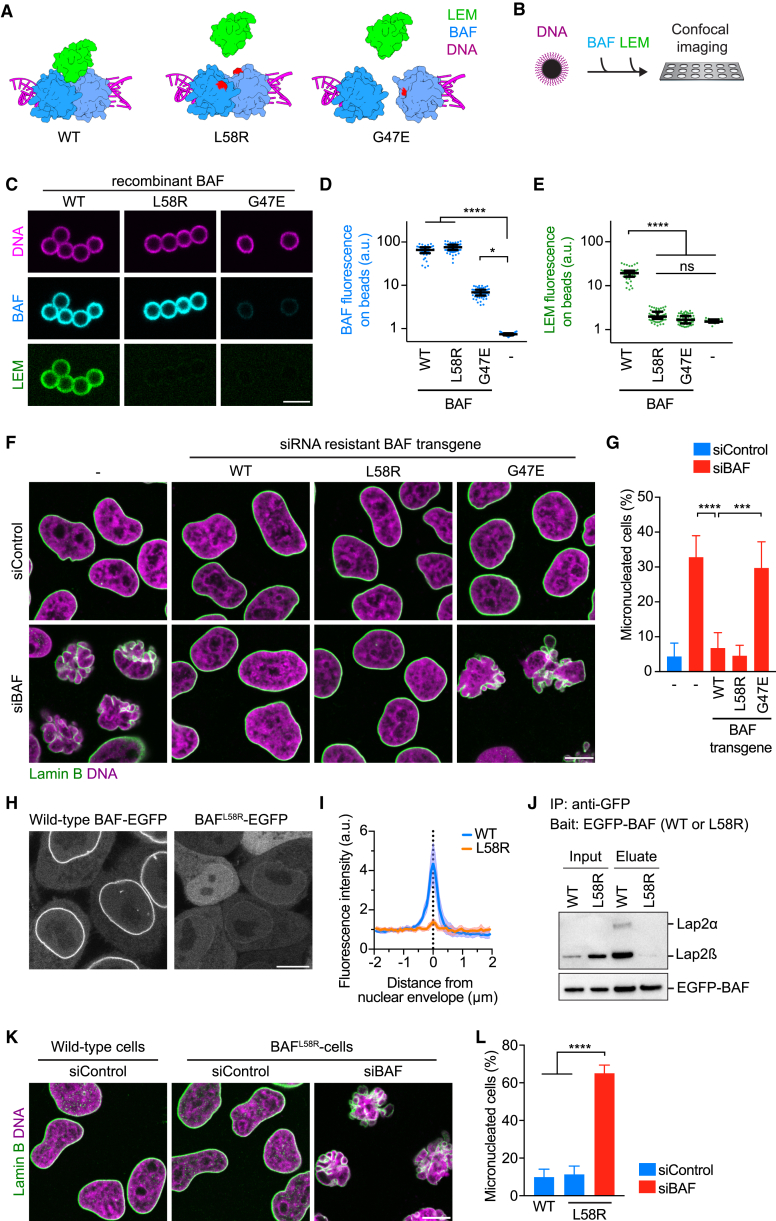
Figure 4.
BAF Dimerization, but Not BAF-LEM Binding, Is Required to Shape a Single Nucleus
(A) Design of point mutations to interfere with BAF-LEM binding or with BAF dimerization. Point mutations (red) are indicated on the structure of the BAF dimer (blue) relative to LEM domain (green). Magenta indicates DNA.
(B) Schematic of the assay.
(C) Analysis of recombinant wild-type (WT) and mutant BAF proteins. Fluorescently labeled recombinant BAF proteins were mixed with DNA-coated beads. Then, fluorescently labeled recombinant LEM domains were added and beads were imaged by confocal microscopy.
(D and E) Quantification of BAF (D) and LEM bound to beads (E) (dots indicate beads, lines indicate median and interquartile range, n ≥ 40 beads per condition).
(F and G) RNAi complementation analysis (F) IF of cells stably expressing siBAF-resistant transgenes. (G) Quantification of micronucleation phenotype. Cells were automatically classified into normal or micronucleated morphology by supervised machine learning (bars indicate mean ± SD, see STAR Methods for sample numbers).
(H) Live localization of BAF-EGFP in wild-type HeLa cells and EGFP-BAFL58R mutant in homozygous L58R mutant HeLa cells.
(I) EGFP fluorescence as in (H) was analyzed along line profiles (curves and range indicate mean ± SD, n = 10 cells per condition).
(J) Immunoprecipitation (IP) of EGFP-tagged BAF as in (H). Co-purified Lap2 was detected by immunoblot analysis using anti-Lap2 and anti-EGFP antibodies.
(K and L) Wild-type or homozygous BAFL58R mutant cells were analyzed by IF (K), and micronucleated cells were automatically quantified by supervised machine learning (L) (bars indicate mean ± SD, see STAR Methods for sample numbers). Scale bar is 5 μm in (C), and 10 μm in (F), (H), and (K).
See also Figure S5.