Figure 3.
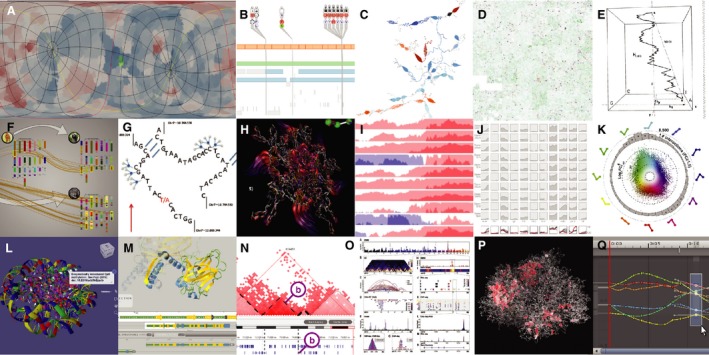
Data abstraction. (A) Flat surface map of green colored docking sites; (B) Transcript focused VariantView; (C) Added granularity in ABySS‐Explorer; (D) Conserved genomic regions in a Hilbert curve; (E) Genomes as 3D H‐curves; (F) Synteny Explorer; (G) Abstraction in Shavit genome browser; (H) Flowlines indicating submolecular motion; (I) Horizon graphs for stacked data; (J) Small multiples as curvemaps; (K) Multistate expression data as rose‐ring; (L) 3D annotation in AutoDesk Molecular viewer; (M) 3D selection from track in Aquaria; (N) Cross‐dimensional section in TADkit; (O) Layout design in Sushi; (P) Chromos VR visualization of chromatin active loops; (Q) Synchronized object and track selection in Unity.
