Figure 2.
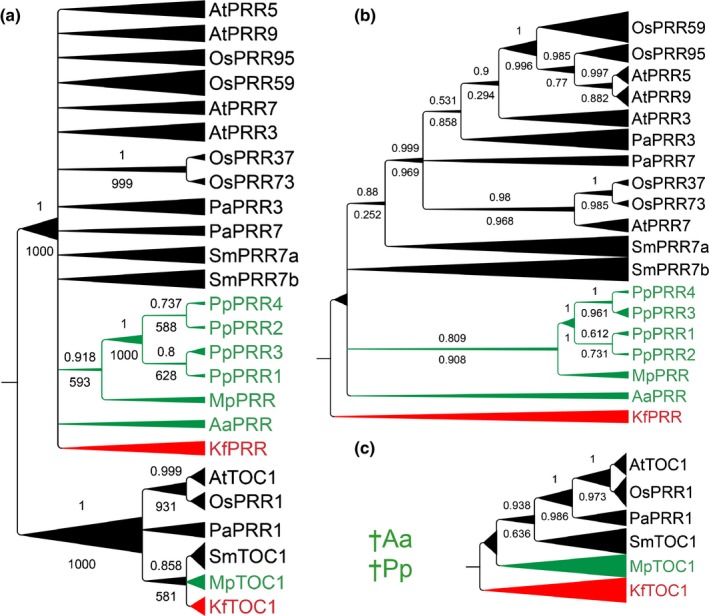
Inferred phylogeny of homologues to the PSEUDO‐RESPONSE REGULATOR (PRR) gene family. Separate trees were constructed from the complete alignment (a), the PRR subfamily (b), and the TOC1 subfamily (c). Trees were constructed using MrBayes and PhyML on an amino acid alignment of proteins retrieved from Arabidopsis thaliana (At), Oryza sativa (Os), Picea abies (Pa), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), Marchantia polymorpha (Mp), Anthoceros agrestis (Aa), and Klebsormidium flaccidum (Kf). The Bayesian tree is shown with posterior probabilities (above) and bootstrap proportions from PhyML analysis (below) for each node. Nodes with conflicting support from the two methods were collapsed. Branch length is relative to the thickness of individual branches: the shortest branches have a straight line and the longest are increasingly triangular. †Aa and †Pp indicate loss of TOC1 in Aa and Pp, respectively (as well as all surveyed hornworts and mosses).
