Figure 1.
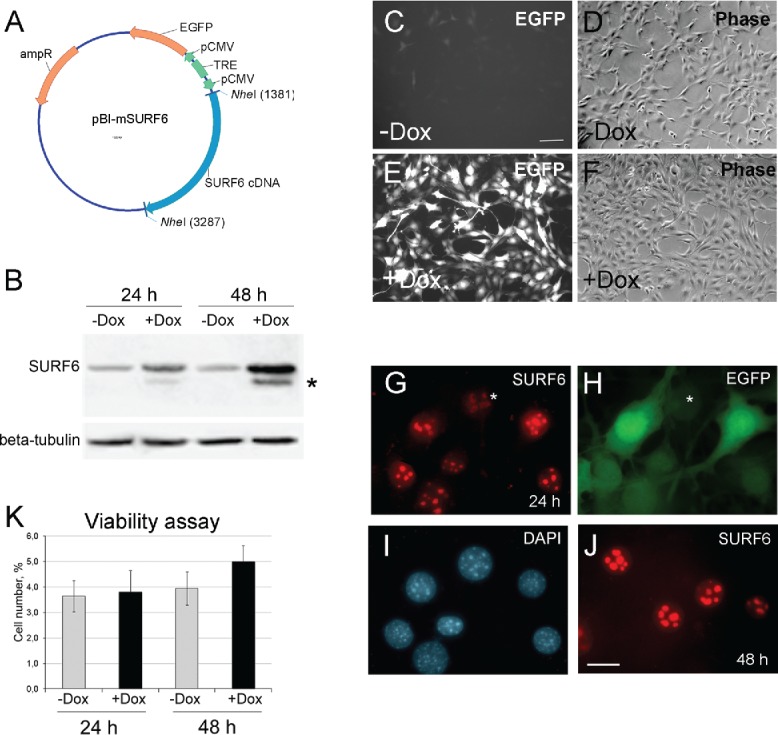
(A) A diagram of the pBI-SURF6 plasmid used for generation of stably transfected mouse NIH/3T3-174 fibroblasts capable to overexpress SURF6 in the presence of doxycycline (Dox). ampR – ampicillin-resistence gene, EGFP – the sequence coding for the EGFP protein, CMV – minimal CMV promoter, TRE – tetracycline-responsive element, SURF6 cDNA – cDNA of the mouse SURF6 gene, Surf6. (B) Immunoblots of control and induced NIH/3T3-174 fibroblasts after incubation without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 and 48 hours. SURF6 was revealed with the anti-SURF6 serum,26 β-tubulin (a loading control) was recognized with a commercial antibody. Star (*) indicates a position of a faster migrating SURF6 form appeared in +Dox fibroblasts. (C-J) General images of NIH/3T3-174 fibroblasts cultured without (-Dox; c, d) or with (+Dox; E-J) 100 ng/ml doxycycline for 48 (C-F and J) or 24 (G-I) hours. (C, E, and H) – EGFP fluorescence, (D and F) – phase contrast, (G and J) – immunolabeling for SURF6; (I) – DAPI staining of chromatin. In the absence of doxycycline, almost no EGFP fluorescence is seen (C). In the presence of doxycycline, the cells are intensely marked for EGFP (E and H), hereby indicating an effectiveness of the pBI-SURF6 plasmid. The phenotype of +Dox fibroblasts (F) remains similar to that of -Dox cells (D). SURF6 is distributed in nucleoli (G and J). A less intense anti-SURF6 immunolabeling of nucleoli is generally seen in cells with a weaker EGFP fluorescence (asterisks, G and H). Large-scale nuclear organization looks similar in different cells (I). Scale bars, (C-F) – 100 µm, (G-J) – 30 µm. (K) Trypan blue excision assay. Bar graphs illustrating the number of dead cells in –Dox (control, grey columns) and +Dox (black columns) cells after 24 and 48 hours of the induction. At each time-point, the control and experimental values do not differ statistically significant (p > 0.05). The data are shown as the mean ± SEM (n = 3).
