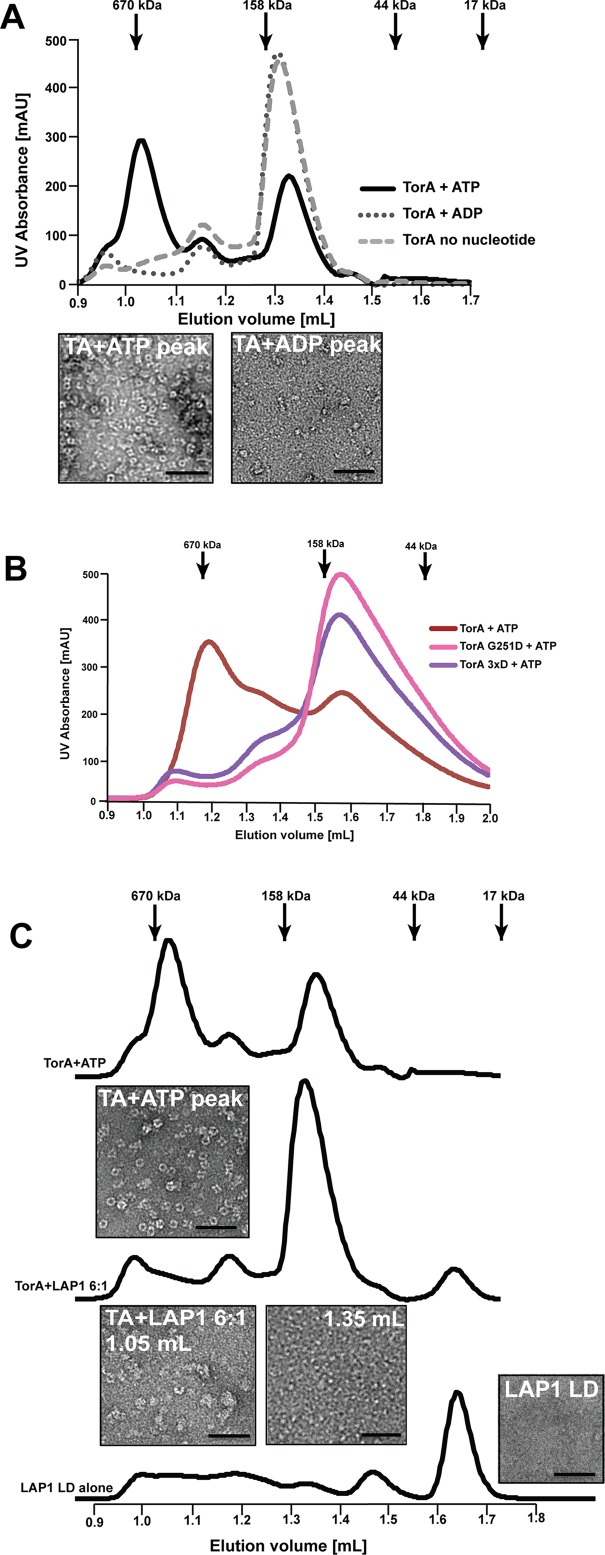
FIGURE 3:
TorsinA forms homo-oligomeric ring structures in the presence of ATP that are disassembled by LAP1LD or mutations of the “back” interface. (A) TorsinA (44 μM) was incubated with 2 mM of ATP or ADP and in the absence of nucleotide and the formation of complexes was assessed by SEC. Fractions were analyzed by negative stain electron microscopy. (B) Purified wild-type, G251D, and 3xD mutant TorsinA (44 μM each) were incubated in the presence of 2 mM ATP and complex formation assessed by SEC. (C) TorsinA (44 μM) was incubated with 2 mM ATP before the addition of 6:1 (mol:mol) Torsin:LAP1LD (5 min. on ice) and complex formation assessed by SEC as before.