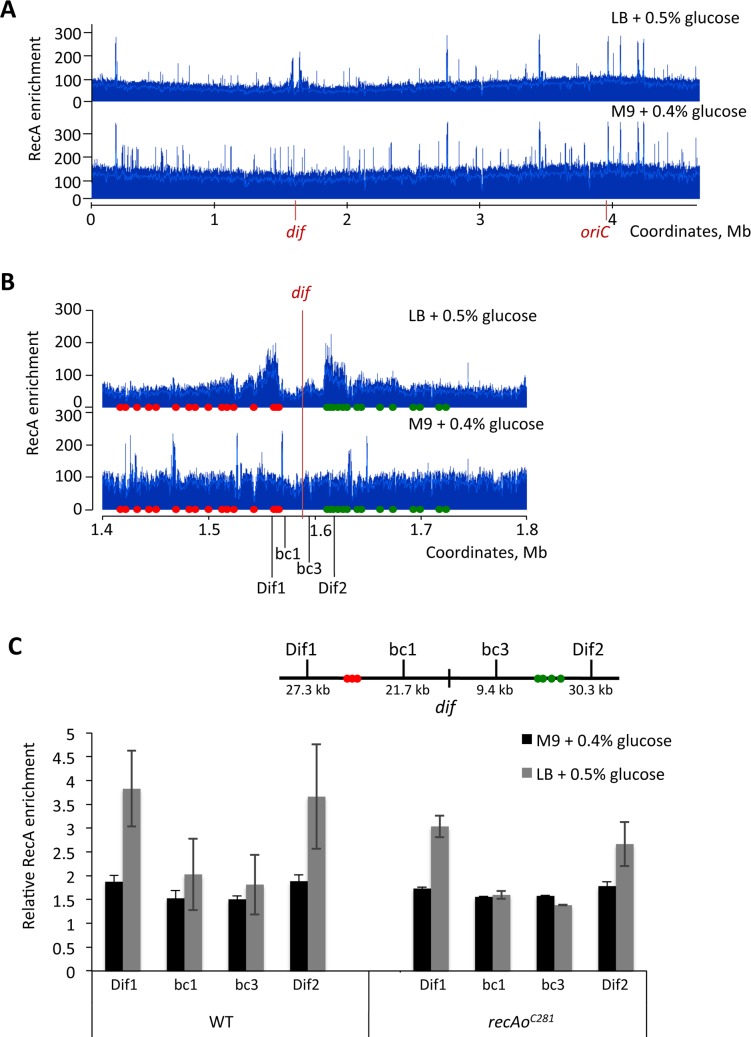
Fig 4. DNA double-strand breaks (DSBs) in the dif/TerC region do not occur in RecB+ cells in minimal medium (M9).
A: Genome-wide RecA binding. RecA-ChIP-seq revealed that the levels of RecA binding at dif/TerC region were not increased in a RecBCD+ strain grown in M9 minimal medium supplemented with 0.4% glucose. The dif site and origin of replication, oriC, are indicated. B: Zoom-in to the terminus region of the chromosome. RecA binding corresponds to the positions of correctly oriented Chi sites. The positions of the correctly oriented Chi sites on each side of the dif site are shown using red (5’-gctggtgg-3’) and green (3’-ccaccagc-5’) circles. Green Chi sites are oriented in such a way that RecBCD recognises them if it moves left to right on the chromosome from a location close to dif. Red Chi sites are recognised by RecBCD that moves in the opposite direction–right to left from a location close to dif. The dif site is located at 1,588 kb. Positions of qPCR primers (Dif1, bc1, bc3 and Dif2) are indicated. C: ChIP-qPCR of wild-type and recAoC281 strains did not show any increase of relative RecA enrichment in the terminus. RecA enrichment in the terminus is normalised to the background level of RecA enrichment at the hycG site. Error bars represent standard error of 3 means. Strains used were wild-type (MG1655) grown in LB medium supplemented with 0.5% glucose and M9 minimal medium supplemented with 0.4% glucose (A, B, C) and recAoC281 grown in LB medium supplemented with 0.5% glucose and M9 minimal medium supplemented with 0.4% glucose (C).