Fig 7. Loss of DNA in the terminus region of recB mutant is independent of DNA sequence and is unaffected by inversion of an adjacent sequence.
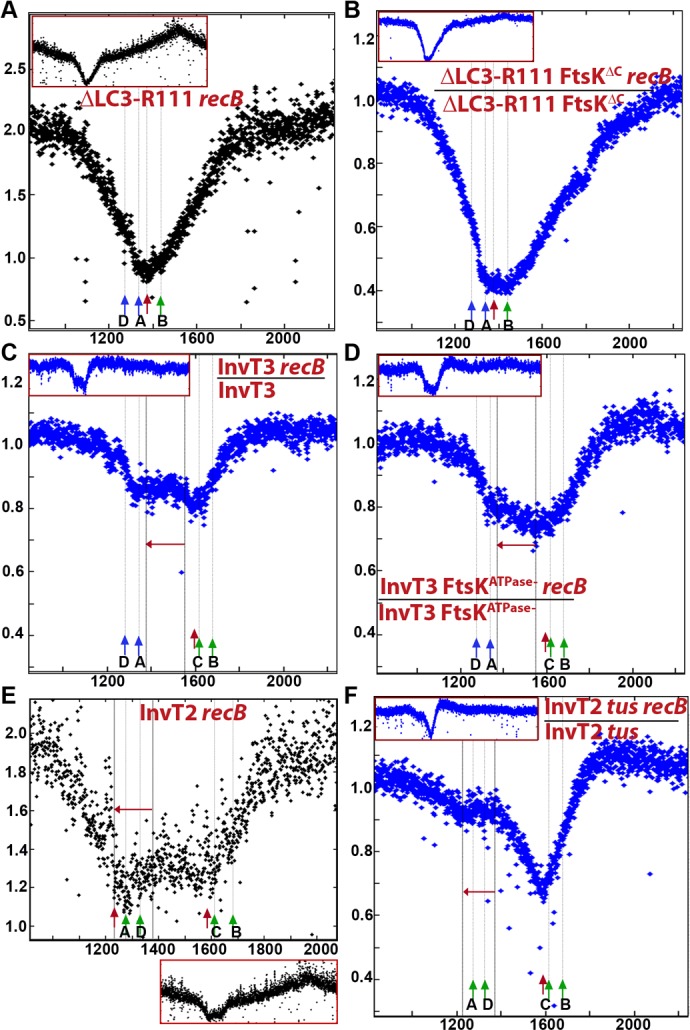
Sequence read frequencies of exponential phase cells normalized to the total amount of reads were calculated for each strain. For the ΔLC3-R111 recB (A) and InvT2 recB (E) mutants, normalized numbers of reads are directly plotted against chromosomal coordinates. For the other mutant strains, ratios of normalized reads in isogenic RecB+ and recB mutants are plotted against chromosomal coordinates. ((B) ΔLC3-R111 ftsKΔCTer, (C) InvT3, (D) InvT3 ftsKATPase, (F) InvT2 tus). Profile ratios of the terminus region are enlarged and profiles of the corresponding entire chromosomes are shown in insets. Original normalized profiles used to calculate ratios are shown in S7, S8 and S9 Figs. The position of dif in InvT2 and InvT3, or of convergence of GC skew in ΔLC3-R111, is indicated by a red arrow. Ter sites that arrest clockwise forks (TerC/TerB in all strains, TerA/TerD in InvT2, green arrow) and counter-clockwise forks (TerA, TerD, except in InvT2, blue arrows) are shown.
