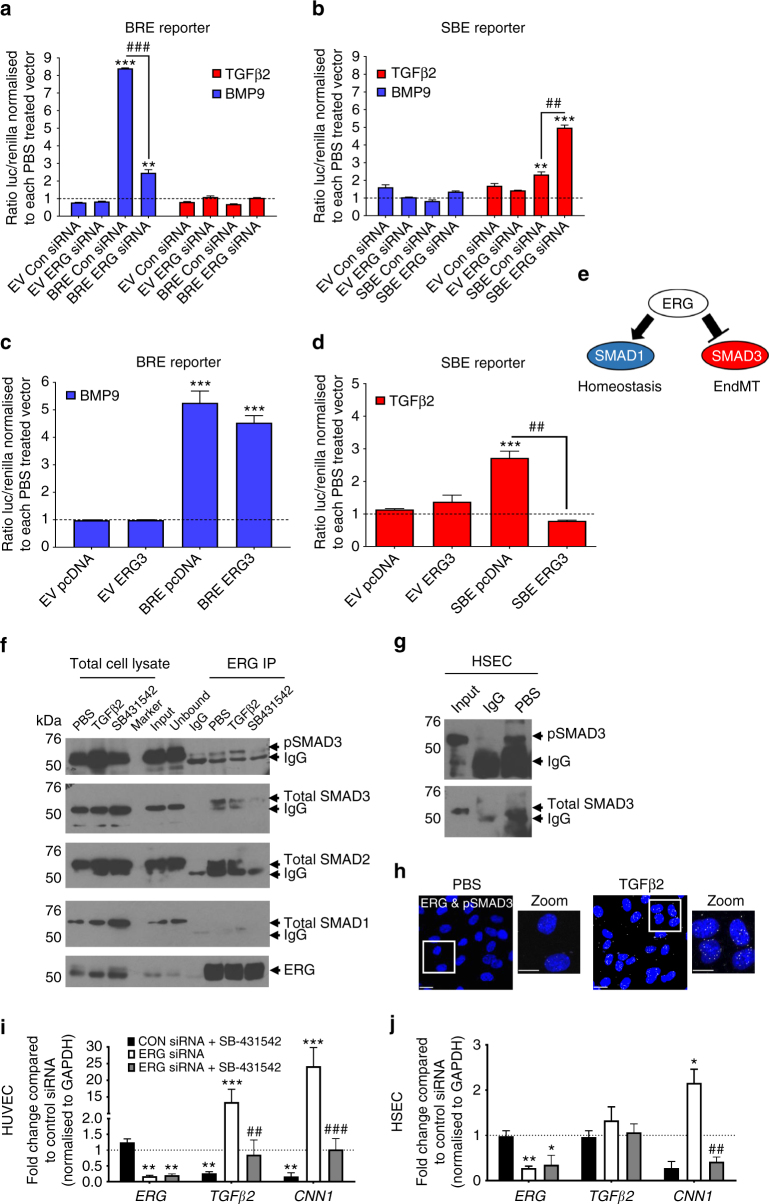
Fig. 2.
ERG differentially regulates transcriptional activity of SMAD1 and SMAD3. ERG-dependent regulation of SMAD1 and SMAD3 activity were assessed by transactivation assays using either SMAD1 reporter pGL3-BRE (BRE reporter, a and c) or SMAD3 reporter pBV-SBE4 (SBE reporter, b and d). HUVEC were either co-transfected with Control siRNA (Con siRNA) or ERG siRNA for 24 h (a and b) or with pcDNA or ERG3 overexpression construct (ERG3) (c and d) prior to treatment with TGFβ2 (red) and BMP9 (blue) or PBS (dashed line) for 18 h (data from pooled HUVEC in triplicate experiments). The ratio of luciferase to renilla from each transfection was normalised to PBS-treated control or to groups co-transfected with (a and b) Con siRNA or (c and d) pcDNA (n = 3). e Schematic of the inverse regulation of SMAD1 and SMAD3 by ERG. f Protein−protein interactions between ERG, SMAD1, SMAD2 and SMAD3 were assessed by Co-IP assay in whole cell lysate from HUVEC. SB-431542 (SB; 10 μM) treatment was performed for 1 h and TGFβ2 (10 ng ml−1) or PBS treatments were performed 30 min prior to lysis. Lysates were immunoprecipitated with mouse IgG or mouse α-ERG and then immuno-blotted for α-ERG, α-SMAD1, α-SMAD2, α-SMAD3 and α-pSMAD3 (Ser423/425). Images representative of four experiments. g ERG-SMAD3 interaction was assessed by Co-IP in untreated HSEC. h Cellular localisation of the interaction between ERG with pSMAD3 was investigated by Proximity Ligation Assay (PLA) following 30 min TGFβ2 treatment in HUVEC (n = 2). i HUVEC or j HSEC were pre-treated with SB-431542 or DMSO prior to transfection with either control siRNA or ERG siRNA for 48 h and analysed by qPCR (n = 3). Data were normalised to GAPDH and compared to control siRNA treated (*) or to ERG siRNA treated with DMSO (#) by unpaired t-test. All graphical data are mean ± s.e.m., * or #P < 0.05, ** or ##P < 0.01, *** or ###P < 0.001