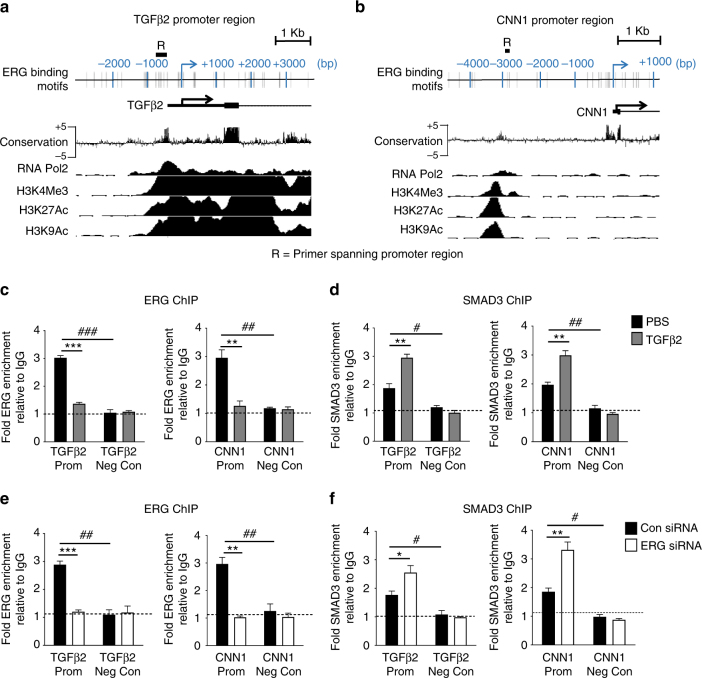
Fig. 3.
ERG is a key factor for endothelial specific inhibition of SMAD3-DNA binding. Putative ERG binding sites (grey bars) are located within the a TGβ2 and b CNN1 promoters upstream of the transcription start site (TSS) (arrow); ENCODE sequence conservation between 100 vertebrates across this region is shown. ENCODE ChIP-seq data profile for H3K4Me3, H3K27Ac, H3K9Ac and RNA polymerase II (RNA Pol2) in HUVEC indicate open chromatin and active transcription. Location of qPCR amplicon region R is indicated. ChIP-qPCR analysis of HUVEC was assessed following treatment with c and d TGFβ2 or PBS for 30 min, or e and f control or ERG siRNA. Chromatin was immunoprecipitated with c and e an α-ERG antibody, d and f α-SMAD3 antibody or control IgG. DNA was analysed by qPCR comparing specific primers against a negative control region for each target. Results are expressed as fold change compared to IgG (n = 3 for all experiments). Basal enrichment was analysed compared to negative control region (#). TGFβ2 or ERG siRNA treatment were compared to PBS treated or control siRNA (*) mean ± s.e.m., * or #P < 0.05, ** or ##P < 0.01, *** or ###P < 0.001 by unpaired t-test