Fig. 8.
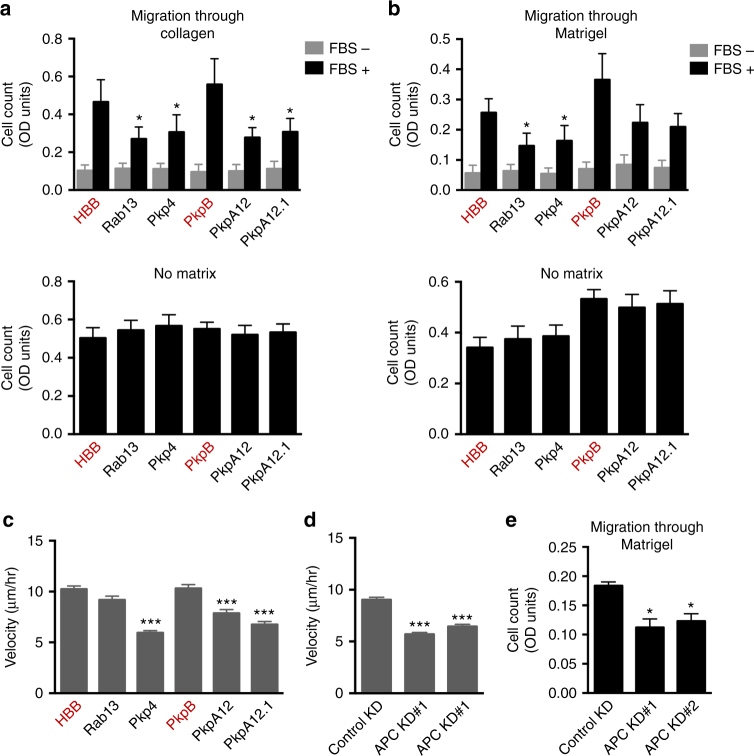
Localization of APC-dependent RNAs is required for efficient migration on 2D and 3D substrates. a, b Cell lines, stably expressing β-globin constructs with the indicated UTRs, were plated on top of collagen gel a or Matrigel b and were induced to migrate by addition of serum (FBS) in the bottom chamber. The amount of cells reaching the bottom was quantified from at least 5 independent experiments, (error bars: standard error). Lower panels show the amount of cells reaching the bottom in the absence of matrix, in parallel experiments, to control for cell viability during the assay. Note that because of the length of the assay, migration in the absence of matrix does not reflect migration speed. c, d Cells stably expressing constructs with the indicated UTRs c or transfected with the indicated siRNAs d, were monitored while randomly migrating. Average velocity values were derived from multiple individual cell tracks in two independent experiments observing 40–70 cells in each. e Migration through Matrigel of control or APC knockdown cells. p-values: ***< 0.0001, *< 0.05 against HBB or si-Control, by analysis of variance with Bonferroni’s multiple comparisons test