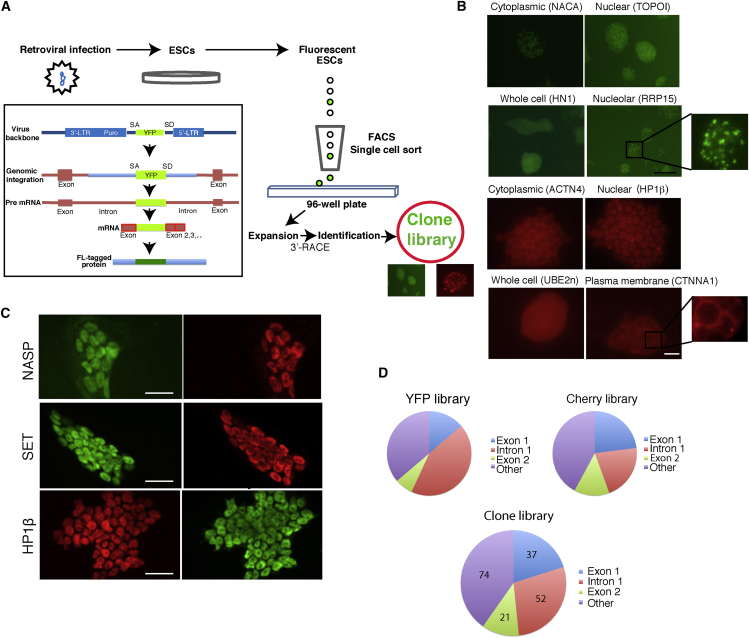
Figure 1.
Clone Library Generation
(A) A schematic representation of the gene-tag clone library preparation. The pBABEpuro CD (Central Dogma)-tagging retroviral vector was used. Fluorescent-tag (FL) labeled ESCs were single sorted using fluorescence-activated cell sorting (FACS) into single wells of 96-well plates. The gene-tagging approach was achieved by the integration of the YFP or Cherry exon (which include strong splice sites but no start/termination codons) inside protein-coding genes, resulting in YFP or Cherry fusion proteins (inset). SA, splice acceptor; SD, splice donor; LTR, long terminal repeat.
(B) Examples, showing different localizations, of endogenously labeled fluorescent fusion proteins: cytoplasmic (NACA); nuclear (TOPOI); whole cell (HN1); and nucleolar (RRP15). Cherry clones (bottom): cytoplasmic (ACTN4); nuclear (HP1β); whole cell (UBE2n); and plasma membrane (CTNNA1). Scale bar, 200 μm.
(C) Endogenously tagged YFP and Cherry fusion proteins are properly localized. Clones expressing YFP-tagged (green, top and middle left) or Cherry-tagged (red, bottom left) proteins were fixed and labeled with the corresponding antibodies (right, Alexa 561-red for the YFP clones and Alexa 488-green for the Cherry clones). From top to bottom: NASP, SET, and HP1β. In all cases perfect co-localizations were observed. Scale bars, 20 μm.
(D) Integration site statistics. The majority of the integration events occurred inside intron 1 and exon 1.