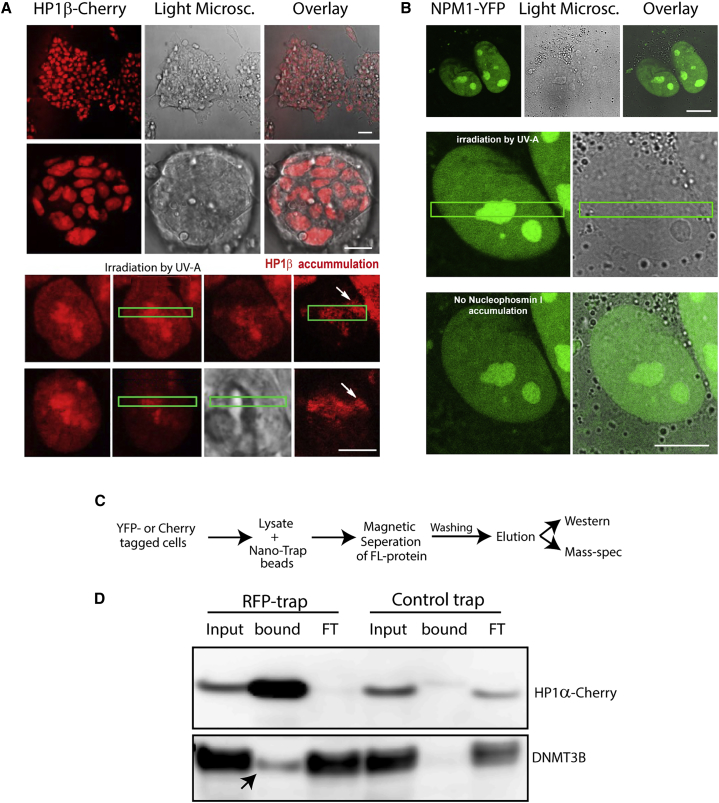
Figure 4.
Functional Validation of Clone Library
(A) Endogenously tagged HP1β fluorescent ESC clones were microirradiated using a 355-nm UVA laser. Cherry accumulation was monitored at the sites of damage. Cells were monitored from 10 s up to 15 min after UVA irradiation. A colony is shown on top (left: fluorescence; middle: bright-field; right: merge), and 1–2 selected cells are shown below. Green boxes depict area irradiated by UV-A. White arrows point at accumulation of HP1β at the DNA damage site. Scale bars (from top to bottom), 50 μm, 10 μm, 4 μm.
(B) Nucleophosmin (NPM1) did not show localization to DNA damage sites. In each panel, a colony is shown on top (left: fluorescence; middle: bright-field; right: merge), and 1–2 selected cells are shown below. Green boxes depict the area irradiated by UV-A. Scale bars (from top to bottom), 4 μm, 2 μm.
(C) Scheme representing the approach used to identify interaction partners combining the clone library with Nano-Trap technology.
(D) Western blots using anti-RFP (top) and anti-DNMT3B (bottom) antibodies showing CoIP of HP1α-Cherry and DNMT3B. RFP trap (lanes 1–3) and Control trap (lanes 4–6) are shown. Arrow indicates the presence of HP1α partner DNMT3B in the bound fraction.