Abstract
Prion-like domains have emerged as important drivers of neurodegenerative disease. Now, Boulay et al. establish that the translocated prion-like domain of the oncogenic EWS-FLI1 fusion protein enables phase-separation events, which inappropriately recruit chromatin-remodeling factors to elicit the aberrant transcriptional programs underlying Ewing’s sarcoma.
Prion-like domains (PrLDs) are found in ~240 human proteins and resemble those that drive prionogenesis of specific yeast proteins. These low-complexity domains possess an unusual amino-acid composition enriched in glycine and uncharged polar amino acids, including glutamine, asparagine, tyrosine, and serine (March et al., 2016). Whether PrLDs enable human proteins to form prions remains unknown. However, PrLDs are found in an emerging cadre of human RNA-binding proteins, including TDP-43, FUS, TAF15, and EWSR1, which are linked to fatal neurodegenerative disorders such as frontotemporal dementia and amyotrophic lateral sclerosis (Couthouis et al., 2012; March et al., 2016). PrLDs render the proteins in which they are housed intrinsically aggregation prone. PrLDs also enable liquid-liquid phase separation, a condensation phenomenon that underpins the biogenesis of diverse membraneless organelles such as para-speckles and stress granules. Liquid-liquid phase separation creates high local concentrations of PrLDs, which can promote nucleation of pathological cross-β fibrils (Lin et al., 2015; March et al., 2016). Similar to the portable nature of yeast prion domains, the ability of PrLDs to promote phase separation and aberrant fibrillization is also portable, and appending PrLDs to various proteins can transfer these properties (Kato et al., 2012; Kwon et al., 2013; Lin et al., 2015). Remarkably, in Ewing’s sarcoma, the second most common pediatric bone cancer, a chromosomal translocation fuses a large portion of the PrLD of EWSR1 to the transcription factor FLI1 (Figure 1) (Tan and Manley, 2009; Toretsky and Wright, 2014). In this issue of Cell, ground-breaking studies by Rivera, Kadoch, and colleagues demonstrate that the PrLD of the EWS-FLI1 fusion protein phase separates and recruits BAF (BRG1/BRM-associated factor, also known as SWI/SNF) chromatin-remodeling complexes to tumor-specific enhancers, thereby activating the transcriptional events of Ewing’s sarcoma. These findings establish that translocated PrLDs can elicit phase-separation events, which unleash aberrant transcriptional cascades that cause cancer (Boulay et al., 2017).
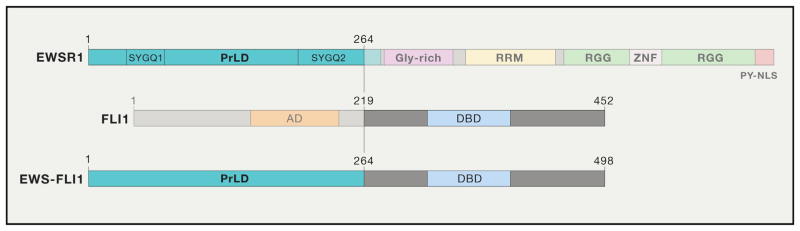
Figure 1. Domain Architecture of EWSR1, FLI1, and EWS-FLI1.
EWSR1 contains an N-terminal prion-like domain (PrLD), a glycine-rich domain (Gly-rich), a RNA-recognition motif (RRM), two RGG domains separated by a zinc-finger domain (ZNF), and a C-terminal PY-nuclear localization sequence (PY-NLS). FLI1 harbors an N-terminal activation domain (AD) and a C-terminal DNA-binding domain (DBD). In the EWS-FLI1 (type 1) fusion protein, amino acids 1–264 of the EWSR1 PrLD are fused with residues 219–452 of FLI1. Note that the SYGQ1 and SYGQ2 portions of the EWSR1 PrLD could substitute for the larger EWSR1 PrLD fragment (residues 1–264) and induce the gene-expression programs of Ewing’s sarcoma when fused to FLI1 (Boulay et al., 2017).
Using an unbiased mass-spectrometry approach, Boulay et al. establish that BAF complexes interact transiently with EWSR1 in a variety of cell types and also with EWS-FLI1 in Ewing’s sarcoma (Boulay et al., 2017). EWS-FLI1 acts as a pioneer factor in mesenchymal stem cells, the probable cell of origin for Ewing’s sarcoma (Riggi et al., 2014). Here, they discover that the pioneer activity of EWS-FLI1 to induce tumor-specific de novo enhancers at GGAA microsatellite repeats is aided by specifically recruiting BAF complexes, which remodel chromatin to spark transcription of EWS-FLI1 target genes. Like the EWS-FLI1 fusion, FLI1 could interact with BAF chromatin-remodeling complexes, but FLI1 could not recruit BAF complexes to tumor-specific de novo enhancers at GGAA microsatellite repeats. Thus, an emergent property provided by the translocated PrLD enables EWS-FLI1 pioneer activity. Moreover, EWS-FLI1 forms numerous nuclear foci, whereas FLI1 remains diffuse (Boulay et al., 2017). Thus, the ability of the PrLD in EWS-FLI1 to phase separate likely enables pioneer activity and recruitment of EWSR1 and BAF complexes to GGAA microsatellite repeats.
The authors further dissect the features of the PrLD that enable EWS-FLI1 to cause Ewing’s sarcoma. More specifically, they establish that, in vitro, purified recombinant EWS-FLI1 and EWSR1 rapidly aggregate, whereas FLI1 remains soluble (Boulay et al., 2017; Couthouis et al., 2012). Thus, the translocated PrLD from EWSR1 renders EWS-FLI1 aggregation prone. Replacement of tyrosine residues with serine in PrLDs alters the molecular grammar of PrLD-mediated phase separation and can reduce fibrillization (Kato et al., 2012; Kwon et al., 2013). Indeed, replacement of 37 tyrosine residues with serine in the EWS-FLI PrLD ablates the ability of EWS-FLI to aggregate in vitro. Importantly, this tyrosine-replaced EWS-FLI1 variant is also unable to bind GGAA microsatellite repeats and to create active enhancers in mesenchymal stem cells. Thus, the ability of the EWS-FLI1 PrLD to phase separate is intimately connected to the ability to activate the transcriptional program of Ewing’s sarcoma. Finally, fusion of short fragments of the EWSR1 PrLD to FLI1 yields proteins that are intrinsically aggregation prone in vitro and phenocopy EWS-FLI1 in inducing the gene-expression program of Ewing’s sarcoma (Figure 1) (Boulay et al., 2017).
The elegant studies of Boulay et al. firmly connect the ability of the EWSR1 PrLD to phase separate to the EWS-FLI1-driven transcriptional program that underpins Ewing’s sarcoma (Boulay et al., 2017). This important work raises several questions. For example, the material properties of the phase-separated EWS-FLI1 that enables the Ewing’s sarcoma transcriptional program remain uncertain. Thus, further studies are needed to clarify whether EWS-FLI1 occupies a liquid phase, a hydrogel phase, or a solid fibril phase to recruit chromatin-remodeling factors and drive transcription. Precisely how the altered phase of EWS-FLI1 recruits BAF also remains uncertain. Previous studies indicate that EWSR1 PrLD fibrils can recruit RNA polymerase II to promote transcription (Kwon et al., 2013), but whether this proposed mechanism operates in Ewing’s sarcoma is unclear. Agents that prevent or reverse phase separation by EWS-FLI1 such as protein disaggregases could have therapeutic utility for Ewing’s sarcoma (March et al., 2016). Phosphorylation of multiple serine and threonine residues in the FUS PrLD promotes the liquid phase and prevents fibrillization (Monahan et al., 2017). Thus, it will be interesting to explore whether increased phosphorylation of the EWS-FLI1 PrLD antagonizes fibrillization and the EWS-FLI1 transcriptional program. Finally, portions of the PrLDs of other human RNA-binding proteins such as FUS and TAF15 also get fused to transcription factors via chromosomal translocations that cause other cancers (Tan and Manley, 2009). For example, a portion of the FUS PrLD gets fused to CHOP in liposarcoma and a portion of the TAF15 PrLD gets fused to Ciz in acute leukemia (Tan and Manley, 2009). It will be enlightening to elucidate whether the translocated FUS and TAF15 PrLDs also must phase separate to drive the aberrant transcriptional programs underlying these cancers.
References
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. Cell. 2017;171(this issue):163–178. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March ZM, King OD, Shorter J. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. EMBO J. 2017 doi: 10.15252/embj.201696394. Published online August 8, 2017. http://dx.doi.org/10.15252/embj.201696394. [DOI] [PMC free article] [PubMed]
- Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB, et al. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]