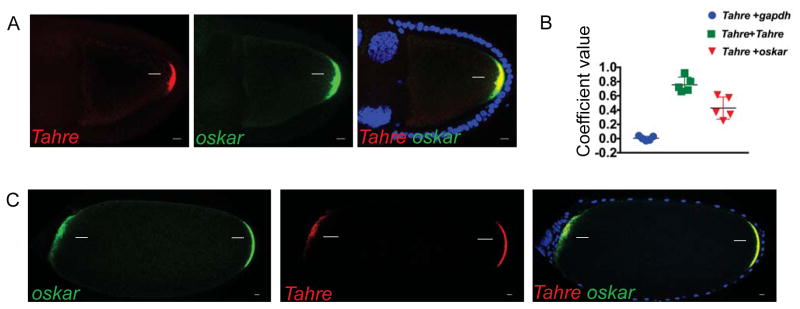
Figure 1. Tahre transposons mimic a germ plasm determinant.
A, confocal images of the simultaneous in situ hybridization of Tahre fluorescently labeled with Quasar 570, oskar 670 and their merge in Drosophila egg chambers of p53−/− ovaries with maximum Z-series projections spanning 5 μm in oocytes at stages 9–10. Scale bars: 10 μm. (Blue) DAPI counterstain. Arrows indicate germ plasm associated signal. (B) Co-localization of Tahre RNAs with oskar RNAs was quantified using automated image analyses. Controls included probes for gapdh and differentially labeled Tahre probes (see STAR Methods). n=5 egg chambers. C, confocal images of in situ co-hybridizations using oskar and Tahre probes (as in A) on stage 14 egg chambers from animals expressing the transgene oskar-bcd3′UTR ovaries in the p53−/− background. Note that Tahre RNAs mirror oskar RNAs, localizing to both the native (rightward arrows) and ecoptic germ plasms (leftward arrows) in these bipolar oocytes. See also Figure S1 and STAR Methods.