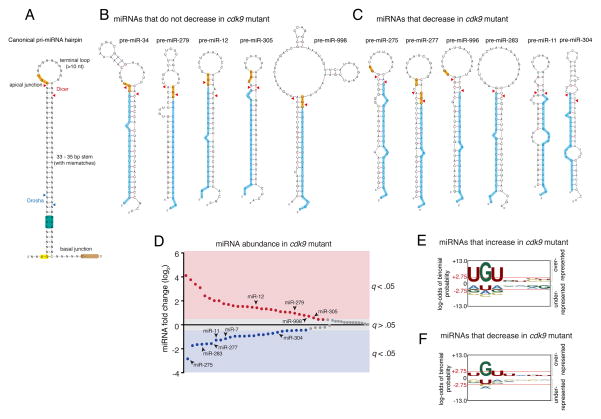
Figure 5. An apical junction sequence is related to Cdk9-dependent processing.
See also Figures S3 and S4. (A) Generalized structural and sequence features of pri-miRNA hairpin. Sequence motifs that affect in vitro processing specificity and efficiency are highlighted. Cleavage sites for Drosha and Dicer are shown with arrowheads. (B) Predicted pre-miRNA structures for those miRNAs whose abundance did not decrease in cdk9 mutants. Highlighted are the UGU motif (yellow) and mature miRNA (blue). (C) Predicted pre-miRNA structures for those miRNAs whose abundance decreased in cdk9 mutants. For B and C, mFold and RNAfold were independently used to predict the pre-miRNA structures. (D) Differential expression of all miRNAs detected by RNA-seq between mutant and wildtype samples. miRNAs are ranked by fold-change, and those whose fold-change is considered significant (a FDR below 5%) are colored. Those miRNAs that are derived from polycistronic genes are noted. (E,F) Logo graphs of nucleotide sequence bias within the apical junctions of pri-miRNA hairpins. Contrasted are the 30 miRNAs whose expression is greater in cdk9 mutants (E) versus the 26 miRNAs whose expression is reduced in cdk9 mutants (F). The y-axes correspond to the binomial probability of residue frequencies, with respect to the background of all Drosophila pre-miRNA sequences. Threshold values of p < 0.05 significance (2.75) are shown in red and marked with red horizontal lines.