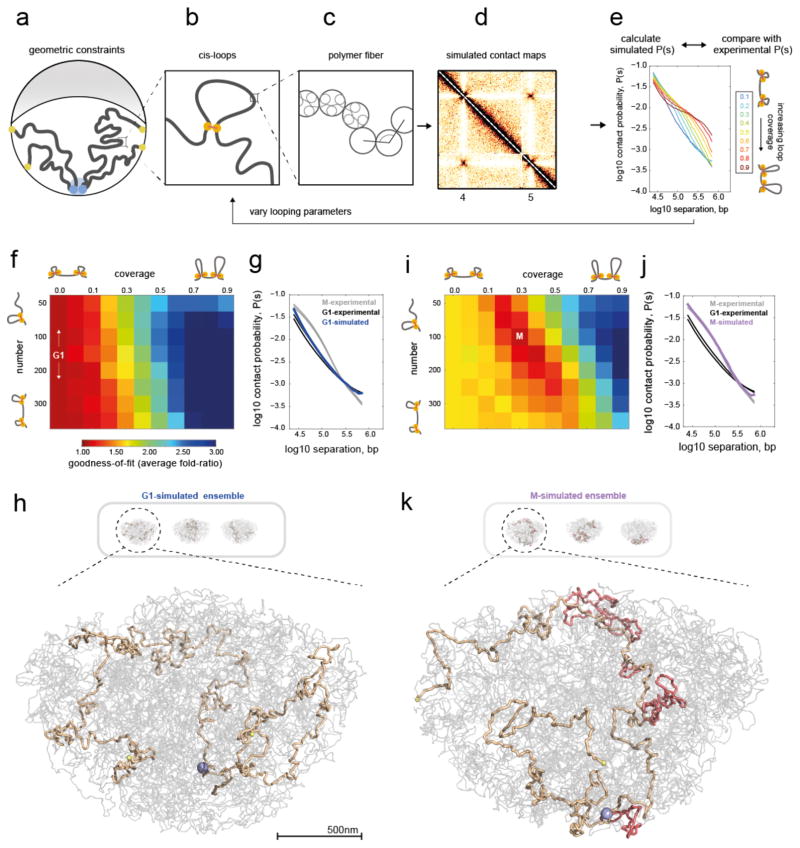
Figure 2. Polymer simulations of the yeast genome support compaction by intra-chromosomal loops in mitosis.
a–e) Overview of simulations.
a) Illustration of geometric constraints used in simulations: confinement to a spherical nucleus, clustering of centromeres (blue), localization of telomeres to the nuclear periphery (yellow), and exclusion of chromatin from the nucleolus (grey crescent).
b) Intra-chromosomal (cis-) loops, generated with a specified coverage and number per yeast genome.
c) Chromatin fiber, simulated as a flexible polymer.
d) Simulated contact maps are generated in simulations with the above constraints.
e) P(s) curves are then calculated from simulated contact maps. Simulations are run with systematically varied cis-loop parameters (coverage and number of loops), and the resulting P(s) curves are compared with experimental data. Shown here are P(s) curves for 150 loops, and a range of coverage.
f) Goodness-of-fit for simulated versus experimental intra-arm P(s) in G1. Goodness-of-fit represents the average fold deviation between simulated and experimental P(s) curves, best-fitting values indicated with white text. The coverage=0.0 column represents the fit for simulations without intra-chromosomal loops.
g) P(s) for best-fitting G1 simulations (coverage=0.0, i.e. no-loops) versus P(s) for each experimental replica of G1 and M.
h) Three sample conformations from ensemble generated in the no-loops simulations; one chromosome highlighted in light brown (from left to right: XI, V, III), with its centromere in blue, telomeres in yellow, and the rest of the genome in grey. Selected conformations show at higher zoom.
i) as F, but for experimental M Hi-C.
j) Best-fitting simulated P(s) for M has N=100–150, coverage=0.3–0.4.
k) Conformations for (N=100, coverage=0.3) with cis-loops additionally highlighted in light red.