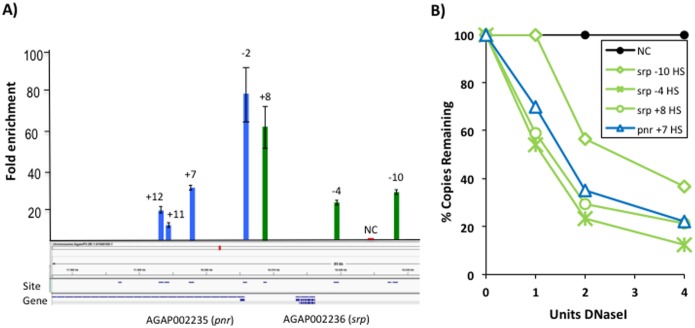
Fig 1. FAIRE-qPCR and DNaseI-qPCR validation of selected cis-regulatory sites detected by FAIRE-seq.
(A) Cis-regulatory sites identified at the locus of A. gambiae’s putative orthologous gene to Drosophila hematopoietic serpent gene were confirmed by FAIRE-qPCR. The cis-regulatory sites identified by FAIRE-seq at -10 Kb, -4 Kb and +8 Kb from A. gambiae’s putative serpent gene (AGAP002236, ensembl!) are enriched in independently prepared open chromatin samples. The cis-regulatory sites identified by FAIRE-seq at -2 Kb, +7 Kb, +11 Kb and +12 Kb from A. gambiae’s putative pannier gene locus (AGAP002235) are also enriched in independently prepared open chromatin samples. (B) Nuclease sensitivity assay by DNaseI-qPCR for some open chromatin sites identified by FAIRE-seq, also detected by FAIRE-qPCR. The genomic coordinates for the tested cis-regulatory sites are relative to the annotated transcriptional start site of each gene.