Figure 1.
Kinetochore Localization and Turnover of Long BubR1 Loop Mutants in HeLa Cells
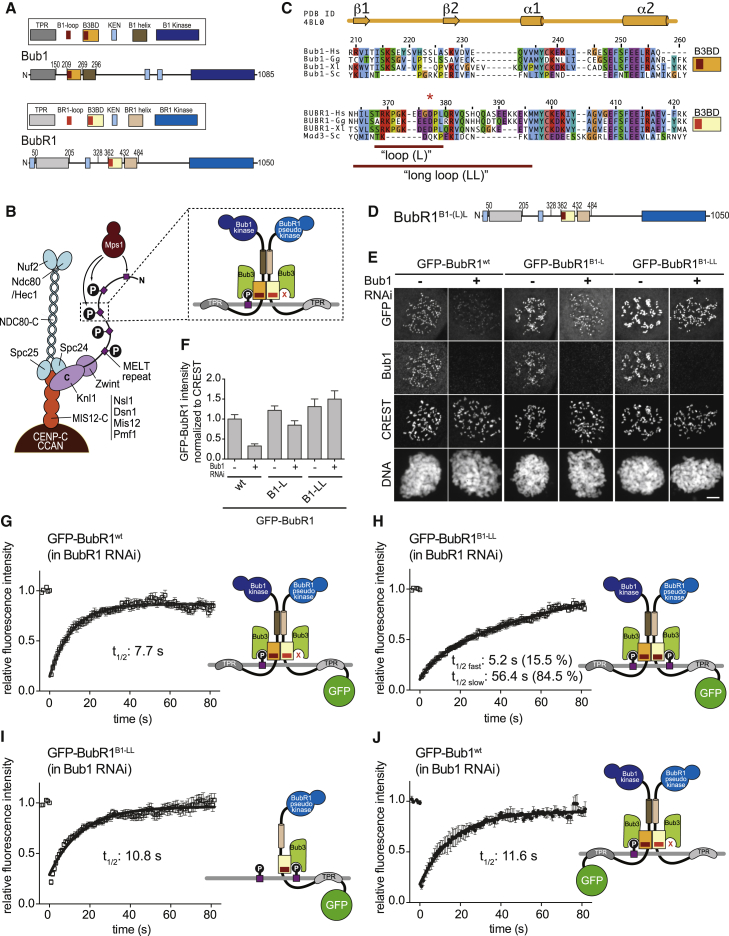
(A) Schematic overview of Bub1 and BubR1 domain organization. B1, Bub1; B3BD, Bub3 binding domain; BR1, BubR1; KEN, lysine-glutamate-asparagine motif; TPR, tetratrico peptide repeat.
(B) Schematic depiction of the outer kinetochore (KMN network). MELT repeats in Knl1 are phosphorylated by the checkpoint kinase Mps1 and recruit Bub1:Bub3. Bub1:Bub3 in turn recruits BubR1:Bub3 via a pseudo-symmetric interaction, which involves equivalent segments of Bub1 and BubR1 comprising the B3BD and the C-terminal extension whose first part is predicted to form a helix in both proteins. The presence of Bub3 on both proteins seems to be essential for this interaction. The TPR regions of human Bub1 and BubR1 bind to non-conserved short motifs of Knl1 named KI1 and KI2, respectively [5, 6, 7].
(C) Multiple sequence alignments of the B3BDs of Bub1 and BubR1 from four different species: Hs, Homo sapiens; Gg, Gallus gallus; Xl, Xenopus laevis; and Sc, Saccharomyces cerevisiae. Mad3 is the budding yeast BubR1 homolog. The initial loop (L) and the long loop (LL) are indicated by the red lines; exact residue numbers are indicated in the main text.
(D) Domain organization of the BubR1 constructs with the Bub1 loop. BubR1B1-L contains Bub1 residues 214–226; BubR1B1-LL contains Bub1 residues 209–235.
(E) Representative images of HeLa cells transfected with the indicated GFP-BubR1 constructs showing that GFP-BubR1B1-LL localizes better to kinetochores than the short loop mutant (B1-L) in presence and absence of endogenous Bub1. In brief, after transfection, cells were depleted of endogenous Bub1 by RNAi, synchronized with a double thymidine block, and arrested in mitosis with nocodazole. The scale bar represents 10 μm.
(F) Quantification of BubR1 kinetochore levels in cells treated as in (E). The graph shows mean intensity from three independent experiments. Error bars represent SEM. Values for BubR1wt in non-depleted cells are set to 1.
(G–J) FRAP analyses of GFP-tagged BubR1wt (G), BubR1B1-LL (H and I), and GFP-Bub1wt in absence of endogenous BubR1 (G and H) or endogenous Bub1 (I and J). Relevant recovery parameters are shown. The graphs show mean with SEM. The cartoons beside the graphs depict the expected mode of kinetochore localization of each construct.
See also Figures S1–S3.