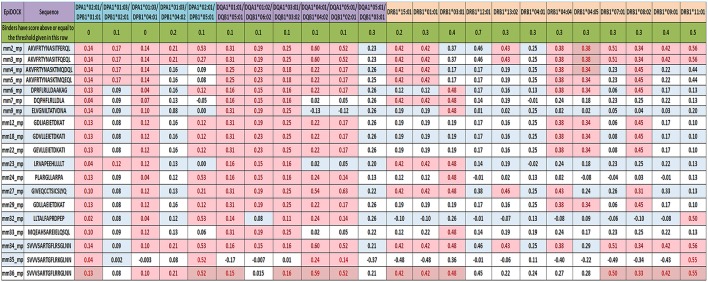
Figure 5.
Reaffirming the promiscuity of MHC class II-binding microbial peptides using structural based predicting tool. EpiDOCK, a homology modeling and molecular docking based algorithmic approach for predicting HLA class II binders were used to further check the promiscuity of all the 19 mimicking peptides. A total of 23 most predominant HLA-DP, HLA-DQ, and HLA-DR alleles were used to predict promiscuity. The input sequences were converted into overlapping fragments of 9-mer before deducing the binding score. The fragment of the peptide that showed maximum affinity was considered to be the representative score of mimicking peptide and was indicated in each row. Binders have scored above or equal to the threshold score [(≥0), (≥0.1), (≥0.2), (≥0.3), (≥0.4), (≥0.5), and (≥0.7)] indicated below each HLA allele and highlighted in red color.