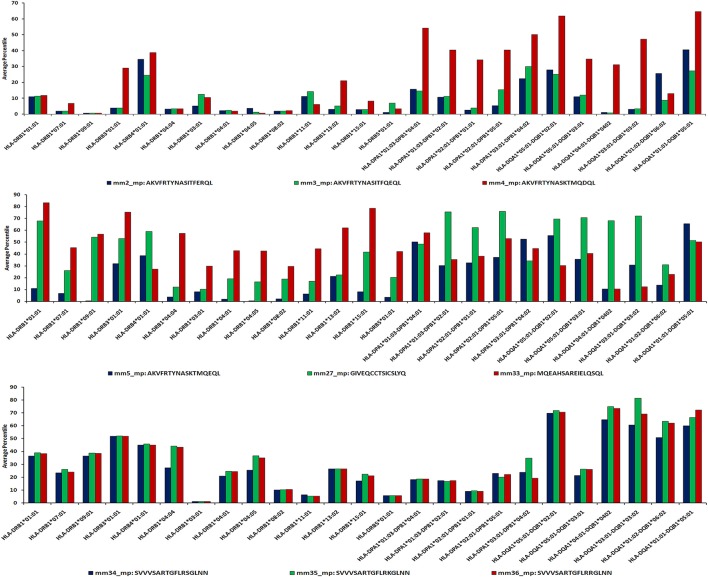
Figure 6.
Sequence-based approach reveals promiscuity of MHC class II-binding microbial peptides. IEDB analysis server allows the prediction of HLA binders using sequence-based algorithmic tool for peptides ≥15 mer. The microbial peptide was fragmented into overlapping 15-mer residues and each fragment was predicted for their binding affinity across MHC class II molecules. The score of each fragment was classified based on predicted IC50 value as strong binders (IC50 ≤ 50), weak binders (50 < IC50 ≤ 500) and non-binders (IC50 > 500). Based on their IC50 values, the peptides are ranked into percentile score for each overlapping fragment. Average percentile score for each fragment represents the overall affinity of the full-length peptide. Low percentile score (≤25) were considered to be HLA class II binders.