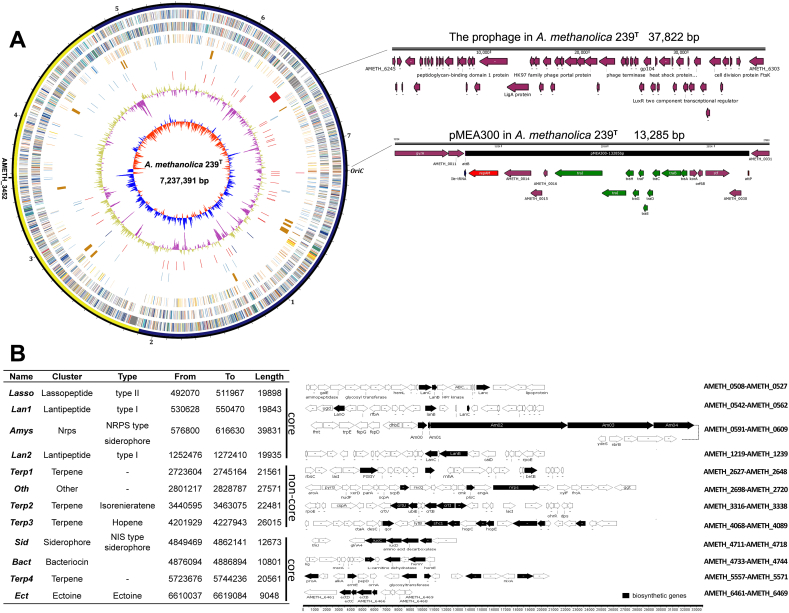
Fig. 1.
Genome atlas of A. methanolica strain 239Twith two characteristic episomes enlarged (A) and the gene clusters encoding biosynthetic functions for specialized metabolites (B). (A) The large circle represents the chromosome: the outer scale is numbered in megabases and indicates the core (blue) and non-core (yellow) regions. The circles are nominated to start from the outside in. The genes in circles 1 and 2 (forward and reverse strands, respectively) are color-coded according to COG functional categories. Circle 3 shows selected essential genes (cell division, replication, transcription, translation, and amino-acid metabolism; the paralogs of essential genes in the non-core regions are not included), circle 4 the phage (red), specialized metabolite clusters (orange) and integrated plasmid pMEA300 (pink), circle 5 the mobile genetic elements (transposases), circle 6 the RNAs, circle 7 the GC content with a calculated ratio of 71.53 mol % and circle 8 the GC bias (blue, values > 0; red, values < 0). The right panel of (A) demonstrates the two characteristic episomes of the A. methanolica genome of strain 239T, i.e., the prophage with its 52 ORFs (upper) and the pMEA300-like integrated plasmid (lower). (B) Gene clusters in the genome of A. methanolica strain 239Tencoding biosynthetic functions for specialized metabolites. The corresponding biosynthesis related genes are dark colored.