FIG 7.
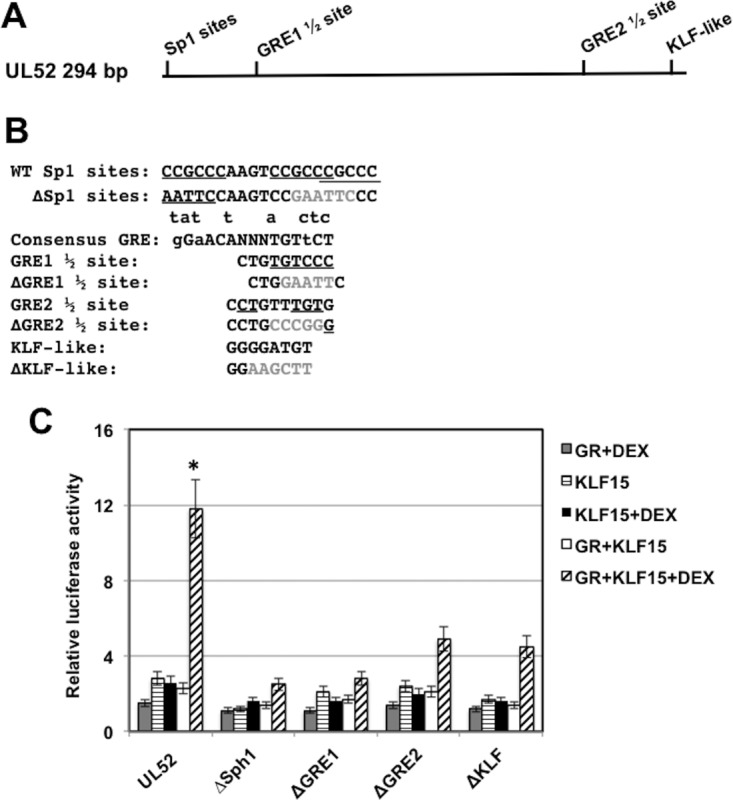
Identification of UL52 intergenic sequences that are important for transactivation by KLF15 and the GR. (A) Location of the GRE half-binding sites (GRE1 1/2 and GRE2 1/2) and potential KLF binding sites in the 294-bp UL52 fragment. (B) UL52 sequences contain three closely linked and partially overlapping Sp1 binding sites that are underlined. The ΔSp1 binding site mutant replaced the Sp1 binding site with an EcoRI restriction enzyme site (GAATTC). The consensus GRE binding site is shown (small letters above consensus are nucleotides that can be part of a functional GRE). The GRE1 half-binding site is shown, and the underlined nucleotides match the consensus. The ΔGRE1 half-binding site contains an EcoRI restriction enzyme site (GAATTC) that replaced the GRE sequences. The GRE2 half-binding site is shown, and the underlined nucleotides match the consensus. The ΔGRE2 half-binding site contains an SmaI restriction enzyme site (CCCGGG) that replaced GRE sequence. The KLF-like binding site is shown, and the ΔKLF mutant contains a HindIII restriction enzyme site (AAGCTT) that replaced the KLF-like binding motif. (C) Neuro-2A cells were cotransfected with the designated plasmid constructs (0.25 μg of DNA) containing the firefly luciferase gene downstream, a plasmid encoding Renilla luciferase (0.05 μg of DNA), KLF15 (0.5 μg DNA), and the GR-expressing plasmid (1 μg of DNA). To maintain equal plasmid amounts in the transfection mixtures, the empty expression vector was added as needed. For these studies, Neuro-2A cells were cultured in 2% stripped fetal calf serum after transfection. Twenty-four hours after transfection the designated Neuro-2A cultures were treated with water-soluble DEX (10 μM; Sigma). At 48 h after transfection, cells were harvested, and protein extracts were subjected to a dual-luciferase assay as described in the Materials and Methods section. The results are the average of three independent experiments, and error bars denote the standard errors. The asterisk denotes a significant difference (P < 0.05) between results with UL52 and the mutant constructs when they are cotransfected with GR plus KLF15 and treated with DEX, as determined by a Student t test.
