Abstract
Introduction
Long noncoding RNAs (lncRNAs) are often dysregulated in cancer tissue and seem to play an important role in neoplastic processes. Recent studies have shown that the HOX transcript antisense intergenic RNA (HOTAIR) may play a role as a marker of prognosis in squamous cell carcinoma of the head and neck (SCCHN). The aim of this study was to perform a meta-analysis of studies focused on the prognostic role of HOTAIR in SCCHN.
Results
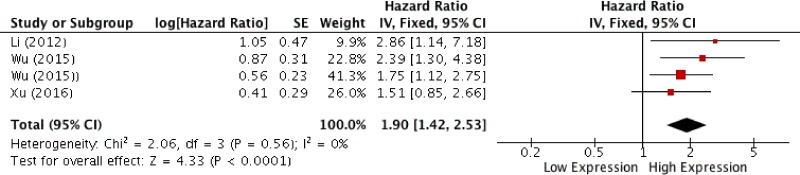
At the end of the selection process, four studies were considered eligible for inclusion in the meta-analysis, comprising a total of 271 patients. Meta-analysis revealed that high expression of HOTAIR was associated with poor overall survival (HR, 1.90; 95% CI: [1.42, 2.53]; p < 0,0001), advanced tumor stage (OR, 3.44; 95% CI: [1.84, 6.43]; p < 0,001) and lymph-node metastasis (OR, 3.31; 95% CI: [1.24, 8.79]; p = 0,02).
Materials and Methods
The literature search was performed in the following databases: PUBMED, SCOPUS, EMBASE and Web of Science, in order to find studies that met the inclusion criteria.
Conclusions
Findings from this systematic review and meta-analysis revealed that HOTAIR represents a potential biomarker of prognosis in patients with squamous cell carcinoma of the head and neck.
Keywords: lncRNA, lncRNAs, non-coding RNA, HOTAIR, biomarker
INTRODUCTION
Squamous cell carcinoma of the head and neck (SCCHN) encompasses a wide and frequent range of neoplasms arising from the epithelium of different sites such as: oral cavity, nasal cavity, oropharynx, hypopharynx and larynx [1, 2]. It has been reported that around 90% of all tumours in this region are squamous cell carcinomas [3]. SCCHNs are among the ten most frequent malignant neoplasms in humans; and due to the high rate of early metastasis to regional cervical lymph-nodes treatment of these patients is a challenge for the clinicians, [4]. It has been estimated that there will be > 300.000 new cases of lip and oral cavity SCC, > 140.000 of oropharyngeal SCC, > 85.000 of nasopharyngeal and > 155.000 of laryngeal SCC annually worldwide, with an increasing incidence and mortality rate [5]. Despite improvement in both surgical and adjuvant therapies, the 5-year survival rate for SCCHN has shown very little improvement over the last decades [6]. For such reasons, searching for biomarkers for diagnosis and prognosis represents a rising hope in order to provide clinicians with new tools for treatment of the disease.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides previously defined as “junk DNA” [7]. They are usually transcribed by RNA polymerase II and can undergo splicing and polyadenylation processes. Although they are not translated into proteins, many of them are linked to some complexes involved in chromatin modification causing an overexpression or silencing of target genes of which most are in cancer pathogenesis [8].
The HOX transcript antisense intergenic RNA (HOTAIR) is one of the most studied lncRNAs [9, 10]. Different studies have focused on the capacity of HOTAIR to cooperate with different chromatin modifying complexes, above all the Polycomb Repressive Complex 2 (PRC2), through its 5′-terminal binding domain [11]. HOTAIR lets PRC2 recognize the target gene, leading to Histone H3 lysine-27 trimethylation, causing a silencing effect. HOTAIR is also able to cooperate with the LSD1 complex and can be regulated by different factors such as miR141, Ago2, c-Myc, TGF-β and small interfering RNAs (siRNAs) [12]. It is also involved in a regulatory control of p53 expression [13]. Different studies suggest that HOTAIR plays an important role in the metastatic process and may be a predictor of poor patient prognosis when highly expressed [14–17].
The aim of this systematic review and meta-analysis was to investigate the link between HOTAIR expression and patient prognosis in SCCHN in order to highlight its potential role as prognostic biomarker.
MATERIALS AND METHODS
Protocol and eligibility criteria
This systematic review and meta-analysis was performed according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines [18] and the Cochrane Handbook [19]. This review has also been registered on the PROSPERO database (registration number: CRD42017057317), before the holding. Studies published in English and fulfilling the following criteria were considered eligible for inclusion in the study: (1) studies focusing on the expression of HOTAIR in SCCHN including more than 50 patients in total, (2) studies showing analysis of correlation between different levels of HOTAIR expression and overall survival, (3) studies reporting a Hazard Ratio (HR) and 95% confidence interval (CI) or Kaplan-Meier curve for HR estimation and (4) studies including quantitative analysis performed through quantitative PCR (qPCR), in situ hybridization (ISH), fluorescence in situ hybridization (FISH), droplet digital PCR (ddPCR) or RNA sequencing data. Accordingly, studies performed on cell lines or animal models, reviews, case reports, overlapping publications and all studies reporting insufficient data for estimation of HR and 95% CI, were excluded. No restriction was applied concerning year of publication.
Information sources and search strategies
The literature search was performed in the following databases: PUBMED, SCOPUS, EMBASE and Web of Science, and independently performed by two of the authors (GT and CC). Mesh terms and free text words were combined through Boolean operators for research on databases. The following terms were used: (“long non-coding RNA” [Mesh] OR “lncRNA” [Mesh] OR “lincRNAs” [Mesh] OR “HOTAIR” [free word] OR “HOX transcript antisense RNA” [Mesh]) AND (“prognosis” [free word] OR “prognostic factor” [Mesh] OR “survival” [Mesh] OR “relapse” [Mesh]) AND ((“head neck” [free word] OR “oral cancer” [Mesh] OR “tongue” [Mesh] OR “pharynx” [Mesh] OR “HNSCC” [free word] OR “OSCC” [free word] OR “tonsil” [Mesh] OR “larynx” [Mesh])). In addition, a direct search on the bibliographies of previously published systematic reviews on the topic was also performed.
Study selection, data collection process and data items
Eligibility assessment of the studies was performed independently in an unblinded standardized manner by two of the authors (GT and CC). In the first round title and abstract was read from the articles found, and studies meeting the inclusion criteria as well as those presenting insufficient data to make a clear decision were then read in full-text. Any disagreement was solved by discussion in a joint session. In order to investigate the level of agreement between the two reviewers, a value of K statistics was calculated. At the end of the selection process papers fulfilling all inclusion criteria were included in the quantitative synthesis. Data extraction was performed through an ad hoc extraction sheet and checked by two authors. In the quantitative synthesis a difference in overall survival was seen for patients with SCCHN showing different levels of HOTAIR expression. Furthermore, differences in: lymph-node metastasis, tumor stage and histological grade were seen between patients with Low or High HOTAIR expression.
Risk of bias assessment, summary measures and planned methods for analyses
Analysis of risk of biases of the included studies was conducted using the Newcastle-Ottawa Scale (NOS) for case control studies. Risk of biases across studies was quantified evaluating the presence of heterogeneity among studies and also investigated through Q and I2 tests. A p-value of Q statistic < 0.05 was considered significant for presence of heterogeneity. Risk of biases across studies was quantified evaluating presence of heterogeneity among studies through Q and I2 tests. The Higgins index was also assessed and classified as follow: < 30% low heterogeneity, 30% to 60% medium heterogeneity, and > 60% high heterogeneity [20]. Overall effects were investigated with a fixed effect model where I2 was less than 50%, if higher, a random effects model was used. Data for overall survival were synthetized as HR and standard error (SE), while for lymph-node metastasis, tumor stage and histological grade the odds ratio (OR) was evaluated. In cases in which HR and its 95% CI was not reported in the article, it was extracted from Kaplan-Meier curves using the method of Tierney et al. [21]. All calculations were performed using Review Manager version 5.2.8 (Cochrane Collaboration, Copenhagen, 153 Denmark; 2014). Results of the meta-analysis were summarized in forest plots, and overall effects were compared using the inverse of variance test. A p-value lower than 0.05 was considered significant for all tests performed in this meta-analysis.
RESULTS
Study selection
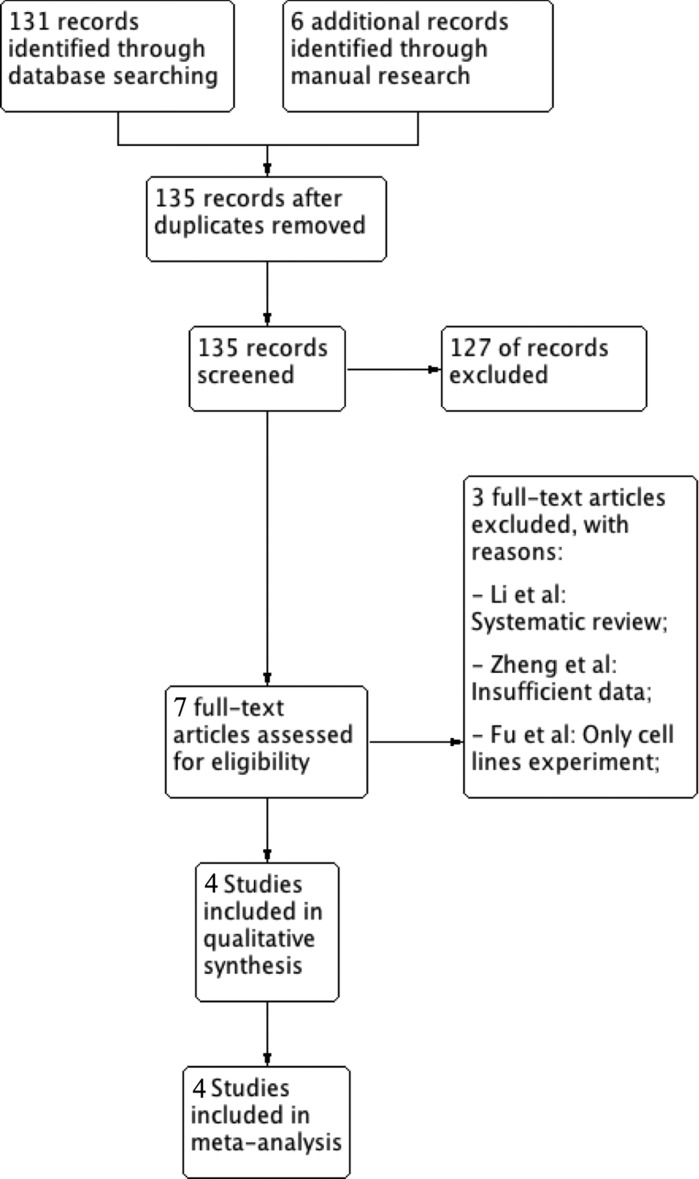
In order to evaluate inclusion in the review, a total of 135 records were screened by title and abstract. Among these only 7 were read full-text and at the end of the selection process 4 articles [22–25] were assessed as eligible for inclusion in the study. Three articles [26–28] were excluded. The reasons for exclusion and the flowchart of the selection process are reported in Figure 1. The value of k-statistic was 0.78 revealing an excellent level of agreement between reviewers.
Figure 1. Flowchart for inclusion of studies in the meta-analysis and reasons for exclusion of articles read in full text.
Study features and risk of bias within studies
In the 4 studies included in the meta-analysis [22–25], a total of 271 samples were analyzed, from tumors in different subsites of the head and neck area (Table 1). All studies were performed in China and published between 2012–2016. Two studies reported data for univariate analysis only [23, 24], while in the other two [22, 25] also multivariate analysis had been performed. In all studies quantification of HOTAIR expression was performed by the use of qPCR. In two studies GADPH was used as a single reference gene [23, 24], another study used 18 rRNA as internal control [25], while in the remaining study authors didn't specify the name of the reference gene used [22]. In addition, all studies reported a follow-up period of at least five years. Risk of bias assessment, performed with the Newcastle-Ottawa scale, showed that all included studies were at low risk of bias (Table 2).
Table 1. Features of the four studies included in the meta-analysis.
| Study | Year | Country | Involved Site | Total number | Detection method | Cut-off | Survival analysis | Multivariate analysis | Follow-up, months |
|---|---|---|---|---|---|---|---|---|---|
| Li | 2012 | China | Larynx | 72 | RT-qPCR | NA | OS | Yes | 60 |
| Wu | 2015 | China | Oral Cavity | 50 | RT-qPCR | Median | OS | No | 60 |
| Wu | 2015 | China | Tongue | 76 | RT-qPCR | NA | OS | No | 100 |
| Xu | 2016 | China | Head and Neck | 73 | RT-qPCR | NA | OS | Yes | 150 |
Table 2. Summary of the risk of bias assessment using the Newcastle-Ottawa scale for case-control studies.
| Author/year | Country | Adequacy of case definition | Representativeness of cases | Controls Selection | Definition of controls | Comparability cases/controls | Ascertainment of exposure | Same method of ascertainment | Nonreponse rate |
|---|---|---|---|---|---|---|---|---|---|
| Li/2012 | China | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | NA |
| Wu/2015 | China | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | NA |
| Wu/2015 | China | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | NA |
| Xu/2016 | China | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | NA |
Synthesis of results and risk of bias across studies
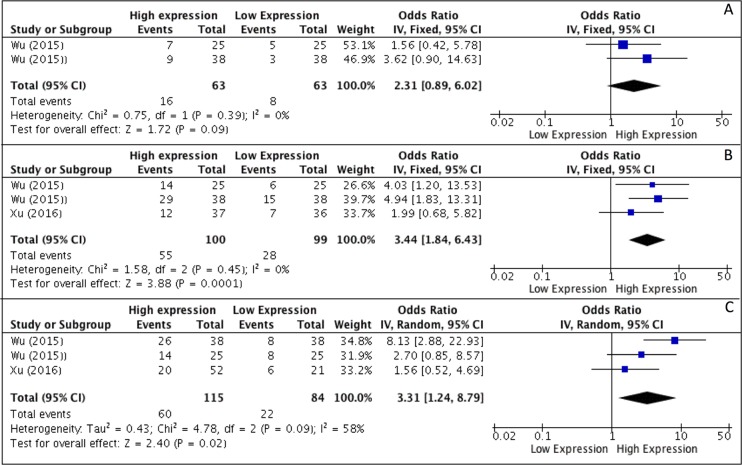
Results of meta-analysis, on the basis of two studies, revealed that higher levels of HOTAIR expression were not associated with higher degree of differentiation (OR, 2.31; 95% CI: [0.89, 6.02]; p = 0,09) (Figure 2A), whereas a correlation to advanced tumor stages was seen (OR, 3.44; 95% CI: [1.84, 6.43]; p < 0,001) (Figure 2B). As heterogeneity was not detected (I2 = 0%) a fixed effects model was used. Analysis of correlations performed with a random effect model (I2 = 58%) revealed a correlation between HOTAIR expression and the rate of LNM (OR, 3.31; 95% CI: [1.24, 8.79]; p = 0,02) (Figure 2C). Analysis of overall survival pooling HRs showed high expression of HOTAIR to be associated with poor OS in patients with SCCHN (HR, 1.90; 95% CI: [1.42, 2.53]; p < 0,0001), (Figure 3). Summary of data extracted from the included studies used for the pooled analysis are shown in Table 3.
Figure 2.
(A–C) Forest plot showing different expression of lncRNA HOTAIR and histological grade (A), tumor stage (B) and lymph-node metastasis (C); the frequency of HOTAIR expression was considered in patients with: Advance Stage (III–IV), positive LNM and Poor/Low Grade of differentiation.
Figure 3. Forest plot for the association between HOTAIR expression and overall survival.
Table 3. Synthesis of data extracted from the included studies related to outcomes pooled in the meta-analysis.
| HOTAIR expression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | High with LNM | Low with LNM | High with high grade | Low with high grade | High with high staging | Low with high staging | High expression | Low expression | HR statistic | Hazard ratios (95% CI) |
| Li | NA | NA | NA | NA | NA | NA | 33 | 39 | Survival curve and data in paper | 2,41–2,856 |
| Wu | 14 | 8 | 7 | 5 | 14 | 6 | 25 | 25 | Survival curve | 2.3 |
| Wu | 26 | 8 | 9 | 3 | 29 | 15 | 38 | 38 | Survival curve | 2.39 |
| Xu | 20 | 6 | NA | NA | 12 | 7 | 52 | 21 | Survival curve | 1.51 |
| Total | 136 | 80 | 24 | 11 | 106 | 54 | 239 | 192 | ||
DISCUSSION
Non-coding RNAs (ncRNAs) are a class of transcripts involved in various cellular processes [29], and classified into three major groups: short ncRNAs (20–50 nt long), medium ncRNAs (50–200 nt long) and long ncRNAs (> 200 nt long). Long non-coding RNAs (lncRNAs) are widely transcribed in the genome and their expression seems to be dysregulated in many diseases [30–32]. Dysregulation of the expression of lncRNAs has been seen in different cancer types, suggesting that these molecules could be involved in tumorigenesis [33–35]. Due to results indicating an important role in cancer, HOTAIR is one of the most studied lncRNAs [11]. HOTAIR is 2.2 kb long and transcribed from the antisense strand of the HOXC gene located on chromosome 2, and consisting of 6 exons [36, 37]. After interaction with the polycomb repressive complex 2 (PRC2) and lysine-specific histone demethylase 1 (LSD1) It is able to regulate gene expression through chromatin dynamics [38, 39]. A recently published study, however, showed that HOTAIR can lead to transcriptional changes independent of PRC2 activity [40]. Furthermore, HOTAIR can act as a competitive endogenous RNA (ceRNA) and regulate levels of some microRNAs [41], for example it can decrease expression of miR-331–3p leading to upregulation of the human epithelial growth factor 2 (HER2), a well-known oncogene [42]. In SCCHN, HOTAIR is able to regulate the activity of the well-known oncogene PTEN, by influencing promoter methylation [25]. In addition, it plays an important role in the epithelial-mesenchymal transition (EMT) repressing expression of E-cadherin through interaction with EZH2 [23, 27]. Overexpression of HOTAIR can also inhibit apoptosis through modification of the mitochondrial membrane and activation of the mitochondrial calcium uptake dependent cell death [43]. It also plays a role in progression and metastasis through a regulatory loop with HuR. In addition, it can work as ceRNA for miR-7 thus avoiding its inhibitory effect on HuR expression [22].
Previous meta-analyses have demonstrated higher expression of HOTAIR to correlate with poor prognosis in patients with cervical [44], ovarian [45], esophageal [44] and gastric [46] cancer. However, to our knowledge no meta-analysis has evaluated its role as a prognostic predictor in SCCHN. In this meta-analysis four studies evaluating correlations between clinical, pathological and survival parameters were included, and a total of 271 patients analyzed. Pooled analysis showed reliable evidence for higher levels of HOTAIR expression to correlate with poor prognosis in SCCHN (HR, 1.90; 95% CI: [1.42, 2.53]; p < 0,0001). In addition, meta-analysis demonstrated a correlation between higher levels of HOTAIR and higher rates of LNM (OR, 3.31; 95% CI: [1.24, 8.79]; p = 0,02). However, presence of heterogeneity (I2= 58%) limits the reliability of these findings, it could be speculated that increasing the number and power of the included studies could render more reliable results. Very interestingly, increased levels of HOTAIR were also associated with advanced tumor stage (OR, 3.44; 95% CI: [1.84, 6.43]; p < 0,001) in the absence of heterogeneity (I2 = 0%). Looking at results for histological degree of differentiation, meta-analysis failed to demonstrate a correlation with HOTAIR expression (OR, 2.31; 95% CI: [0.89, 6.02] p = 0.09). However, only two studies were pooled in the evaluation of this outcome, resulting in very low power of evidences.
Taken together, the present results suggest a potentially important role for HOTAIR as a biomarker of aggressiveness in SCCHN. Results of this meta-analysis should, however, be read with caution due to some obvious limitations. First of all, the number of studies included was low. Secondly a geographical bias may be present as all studies were performed in China, rendering data only from one ethnic group, which could influence the validity of the results. As demonstrated in previous studies, people of different race/ethnicity vary in their risk of developing SCCHN [47, 48]. In addition, differences in prognosis have been seen for patients of different ethnicities, linked both to the genetic and the lifestyle variabilities [49]. As it is known that the frequency of smoking or chewing tobacco, alcohol consumption and HPV infection differs among populations, there can be a “geographical bias” between studies [48, 50]. In addition, the studies included in the meta-analysis have evaluated samples from different subsites within the head and neck area, sites that can differ in both clinical and molecular behavior [51–53]. Furthermore, not all studies reported time-to-event outcome with a multivariate approach, and cut-off values differed and were, in some of the included articles, not even reported. Multivariate analysis allows adjustment for patient-related factors, known as covariates or confounders, which could potentially affect the survival time of the patients [54, 55]. Whether for example a higher number of patients with advanced stage is present in a group, the difference in prognosis between groups could be influenced by the Staging distribution and not HOTAIR expression. On the other hand, the low rate of heterogeneity recorded in three of the studies, combined with a follow-up time of at least 60 months enhances the reliability of findings.
In conclusion, findings from this meta-analysis indicate that higher expression of lncRNA HOTAIR is associated with poor prognosis, advanced tumor stage and higher rate of lymph-node metastasis.
Based on the present results, quantification of HOTAIR could be recommended as a complement associated with evaluation of other prognostical, clinical and molecular biomarkers, in order to categorize patients with high risk of death and thus to help clinicians in the choice of best personalized treatment option. Furthermore, this study encourages the execution of large and well-standardized multicenter studies on human samples of different ethnicity in order to confirm these preliminary findings.
ACKNOWLEDGMENTS AND FUNDING
This study was supported by grants from the Cancer Research Foundation in Northern Sweden, the Swedish Cancer Society Contract number 16 03 36, Västerbotten County Council and Umeå University.
Footnotes
CONFLICTS OF INTEREST
Authors declare to not have any type conflicts of interest related to this study.
REFERENCES
- 1.Arfe A, Malvezzi M, Bertuccio P, Decarli A, La Vecchia C, Negri E. Cancer mortality trend analysis in Italy, 1970–2007. Eur J Cancer Prev. 2011;20:364–374. doi: 10.1097/CEJ.0b013e328345f99e. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Arfe A, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2011. Ann Oncol. 2011;22:947–956. doi: 10.1093/annonc/mdq774. [DOI] [PubMed] [Google Scholar]
- 3.Dobrossy L. Epidemiology of head and neck cancer: magnitude of the problem. Cancer Metastasis Rev. 2005;24:9–17. doi: 10.1007/s10555-005-5044-4. [DOI] [PubMed] [Google Scholar]
- 4.Abu-Ghanem S, Yehuda M, Carmel NN, Leshno M, Abergel A, Gutfeld O, Fliss DM. Elective Neck Dissection vs Observation in Early-Stage Squamous Cell Carcinoma of the Oral Tongue With No Clinically Apparent Lymph Node Metastasis in the Neck: A Systematic Review and Meta-analysis. JAMA Otolaryngol Head Neck Surg. 2016;142:857–865. doi: 10.1001/jamaoto.2016.1281. [DOI] [PubMed] [Google Scholar]
- 5.Gupta B, Johnson NW, Kumar N. Global Epidemiology of Head, Neck Cancers: A Continuing Challenge. Oncology. 2016;91:13–23. doi: 10.1159/000446117. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 7.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–10996. doi: 10.18632/oncotarget.2770. http://doi.org/10.18632/oncotarget.2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014;46:1011–1015. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 11.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai N, Xia Y, Yin R, Liu J, Gao F. A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. Onco Targets Ther. 2016;9:5713–5720. doi: 10.2147/OTT.S110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao X, Ren T, Huang Y, Sun K, Wang S, Liu K, Zheng B, Guo W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. BioMed Res Int. 2013;2013:251098. doi: 10.1155/2013/251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Liu J, Zheng Y, You L, Kuang D, Liu T. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour Biol. 2014;35:11887–11894. doi: 10.1007/s13277-014-2453-4. [DOI] [PubMed] [Google Scholar]
- 17.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62:e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions. Chichester, England; Hoboken, NJ: Wiley-Blackwell; 2008. The Cochrane Collaboration. [Google Scholar]
- 20.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 21.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu CZ, Jiang C, Wu Q, Liu L, Yan X, Shi R. A Feed-Forward Regulatory Loop between HuR and the Long Noncoding RNA HOTAIR Promotes Head and Neck Squamous Cell Carcinoma Progression and Metastasis. Cell Physiol Biochem. 2016;40:1039–1051. doi: 10.1159/000453160. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Liu T, Wang X, Zhou X, Yu J, Hao X. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Xie H. Expression of long noncoding RNA-HOX transcript antisense intergenic RNA in oral squamous cell carcinoma and effect on cell growth. Tumour Biol. 2015;36:8573–8578. doi: 10.1007/s13277-015-3598-5. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Cao Y, Gong X, Li H. Long noncoding RNAs in head and neck cancer. Oncotarget. 2017;8:10726–10740. doi: 10.18632/oncotarget.12960. http://doi.org/10.18632/oncotarget.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Xiao X, Wu C, Huang J, Zhang Y, Xie M, Zhang M, Zhou L. The role of long non-coding RNA HOTAIR in the progression and development of laryngeal squamous cell carcinoma interacting with EZH2. Acta Otolaryngol. 2017;137:90–98. doi: 10.1080/00016489.2016.1214982. [DOI] [PubMed] [Google Scholar]
- 28.Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang HD, Li G, Zhang JF. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7:4712–4723. doi: 10.18632/oncotarget.6731. http://doi.org/10.18632/oncotarget.6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar C, Chatterjee S, Thum T. Long Noncoding RNAs in Cardiovascular Pathology, Diagnosis, and Therapy. Circulation. 2016;134:1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 31.Majidinia M, Mihanfar A, Rahbarghazi R, Nourazarian A, Bagca B, Avci CB. The roles of non-coding RNAs in Parkinson's disease. Mol Biol Rep. 2016;43:1193–1204. doi: 10.1007/s11033-016-4054-3. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 34.Gan L, Xu M, Zhang Y, Zhang X, Guo W. Focusing on long noncoding RNA dysregulation in gastric cancer. Tumour Biol. 2015;36:129–141. doi: 10.1007/s13277-014-2894-9. [DOI] [PubMed] [Google Scholar]
- 35.Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: where do we go from here? Int J Mol Sci. 2015;16:1395–1405. doi: 10.3390/ijms16011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Wang Z, Shi H, Li H, Li L, Fang R, Cai X, Liu B, Zhang X, Ye L. HBXIP and LSD1 Scaffolded by lncRNA Hotair Mediate Transcriptional Activation by c-Myc. Cancer Res. 2016;76:293–304. doi: 10.1158/0008-5472.CAN-14-3607. [DOI] [PubMed] [Google Scholar]
- 40.Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B, Margueron R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017;36:981–994. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Zhou X, Wu Y, Wang Y, Chen L, Li P, Liu S, Sun S, Ren Y, Mei M, Wang X, Zhang L. Targeting HOTAIR Induces Mitochondria Related Apoptosis and Inhibits Tumor Growth in Head and Neck Squamous Cell Carcinoma in vitro and in vivo. Curr Mol Med. 2015;15:952–960. doi: 10.2174/1566524016666151123112716. [DOI] [PubMed] [Google Scholar]
- 44.Liu S, Zhang M, Qu P. Expression level and clinical significance of HOX transcript antisense intergenic RNA in cervical cancer: a meta-analysis. Sci Rep. 2016;6:38047. doi: 10.1038/srep38047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo P, Liu XF, Wang YC, Li ND, Liao SJ, Yu MX, Liang CZ, Tu JC. Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: a systematic review and meta-analysis. Oncotarget. 2017;8:23927–23936. doi: 10.18632/oncotarget.14760. http://doi.org/10.18632/oncotarget.14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu FT, Qiu C, Luo HL, Zhang Y, Xia GF, Hao TF, Zhu PQ. The association of HOTAIR expression with clinicopathological features and prognosis in gastric cancer patients. Panminerva Med. 2016;58:167–174. [PubMed] [Google Scholar]
- 47.Weatherspoon DJ, Chattopadhyay A, Boroumand S, Garcia I. Oral cavity and oropharyngeal cancer incidence trends and disparities in the United States: 2000–2010. Cancer Epidemiol. 2015;39:497–504. doi: 10.1016/j.canep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Kumar SK, Sedghizadeh PP, Jayakar AN, Shuler CF. Oral squamous cell carcinoma incidence by subsite among diverse racial and ethnic populations in California. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;105:470–480. doi: 10.1016/j.tripleo.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Osazuwa-Peters N, Massa ST, Christopher KM, Walker RJ, Varvares MA. Race and sex disparities in long-term survival of oral and oropharyngeal cancer in the United States. J Cancer Res Clin Oncol. 2016;142:521–528. doi: 10.1007/s00432-015-2061-8. [DOI] [PubMed] [Google Scholar]
- 50.Zain RB. Cultural and dietary risk factors of oral cancer and precancer—a brief overview. Oral Oncol. 2001;37:205–210. doi: 10.1016/s1368-8375(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 51.Balasubramanian D, Ebrahimi A, Gupta R, Gao K, Elliott M, Palme CE, Clark JR. Tumour thickness as a predictor of nodal metastases in oral cancer: comparison between tongue and floor of mouth subsites. Oral Oncol. 2014;50:1165–1168. doi: 10.1016/j.oraloncology.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Green VL, Michno A, Stafford ND, Greenman J. Increased prevalence of tumour infiltrating immune cells in oropharyngeal tumours in comparison to other subsites: relationship to peripheral immunity. Cancer Immunol Immunother. 2013;62:863–873. doi: 10.1007/s00262-013-1395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boldrup L, Coates PJ, Wahlgren M, Laurell G, Nylander K. Subsite-based alterations in miR-21, miR-125b, and miR-203 in squamous cell carcinoma of the oral cavity and correlation to important target proteins. J Carcinog. 2012;11:18. doi: 10.4103/1477-3163.104007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark TG, Bradburn MJ, Love SB, Altman DG. Survival analysis part I: basic concepts and first analyses. Br J Cancer. 2003;89:232–238. doi: 10.1038/sj.bjc.6601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradburn MJ, Clark TG, Love SB, Altman DG. Survival analysis part II: multivariate data analysis—an introduction to concepts and methods. Br J Cancer. 2003;89:431–436. doi: 10.1038/sj.bjc.6601119. [DOI] [PMC free article] [PubMed] [Google Scholar]