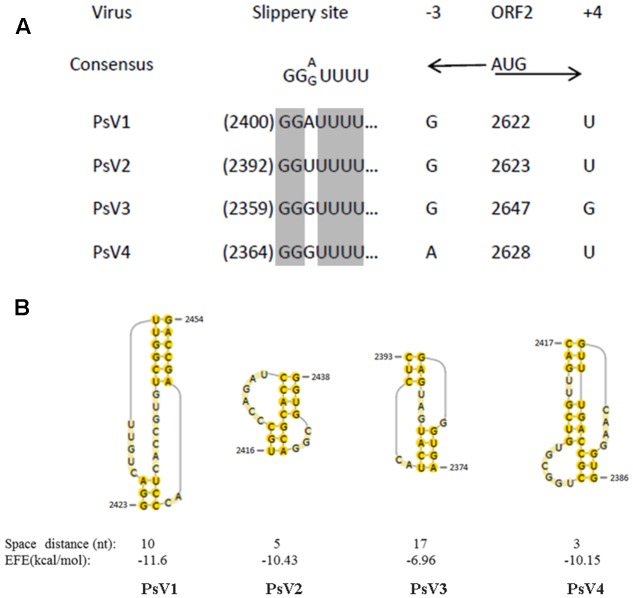
FIGURE 4.
Putative slippery sites and pseudoknot structures predicted in PsVs. (A) The putative slippery sequences (XXXYYYZ) involved in -1 translational frameshift. The PsVs nucleotide sequences at –3 and +4 relative to the AUG start codon are indicated by black arrows. (B) The predicted pseudoknot structures downstream of the potential slippery sequences for –1 frameshifting are shown by yellow shadow. Spacer distance represents the number of nucleotides between the slippery site and the pseudoknot. EFE (kcal/mol) indicates the minimal free energy.