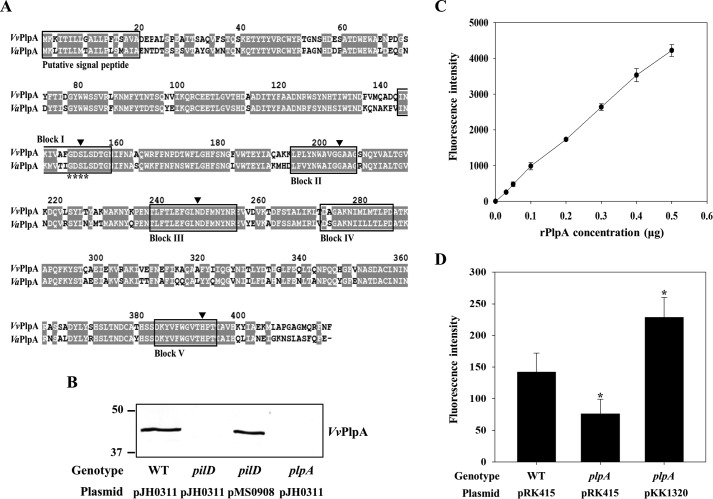
Figure 1.
Analysis of amino acid sequence, secretion, and enzymatic activity of VvPlpA. A, the amino acid sequences retrieved from the NCBI protein database (http://www.ncbi.nlm.nih.gov) (accession numbers ADV88279 for VvPlpA and AAY26144 for VaPlpA) were aligned using the Clustal Omega and Genedoc program. Identical residues (gray boxes) and missing sequence (dash) are indicated. The putative signal peptides and consensus blocks for SGNH hydrolases are boxed by a black line. The GDSL motif is indicated by asterisks. The four residues conserved in the SGNH hydrolases are indicated by black-filled inverse triangles. B, VvPlpA in the supernatants of the V. vulnificus strains grown to an A600 of 0.9 was detected by Western blot analysis. Molecular size markers (Bio-Rad) are shown in kDa. C and D, PLA2 activity was examined by measuring fluorescence intensity emitted from the fluorogenic phosphatidylcholine substrate (red/green BODIPY PC-A2) after incubation with various amounts of the rPlpA (C) or supernatants of the V. vulnificus strains (D) for 0.5 h. Error bars, S.D. Statistical significance was determined by Student's t test. *, p < 0.05 relative to the wild type. WT pJH0311 and WT pRK415, wild type; pilD pJH0311, pilD mutant; plpA pJH0311 and plpA pRK415, plpA mutant; pilD pMS0908 and plpA pKK1320, complemented strains.