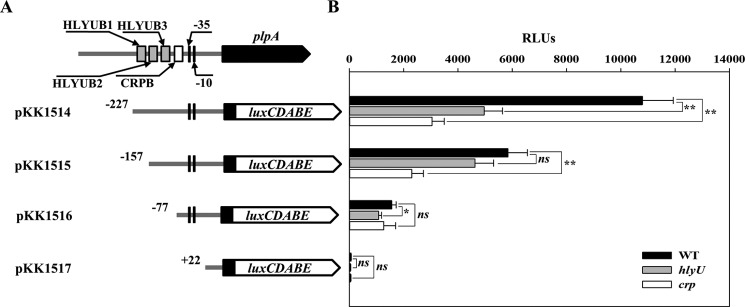
Figure 7.
Deletion analysis of the PplpA regulatory region. A, construction of plpA-lux fusion pKK reporters. PCR fragments carrying the plpA-regulatory region with 5′-end deletions were subcloned into pBBR-lux (64) to create each pKK reporter. Solid lines, the upstream region of plpA; black blocks, the plpA coding region; white blocks, luxCDABE. The wild-type plpA-regulatory region is shown on top with the proposed −10 and −35 regions, and the binding sites for HlyU (HLYUB1, HLYUB2, and HLYUB3; gray boxes) and CRP (CRPB; white box) were determined later in this study (Fig. 8, C and D). B, cellular luminescence was determined from the wild type (black bars), hlyU mutant (gray bars), and crp mutant (white bars) containing each pKK reporter as indicated. Cultures grown to an A600 of 0.9 were used to measure the cellular luminescence. Error bars, S.D. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.005; ns, not significant. WT, wild type; hlyU, hlyU mutant; crp, crp mutant.