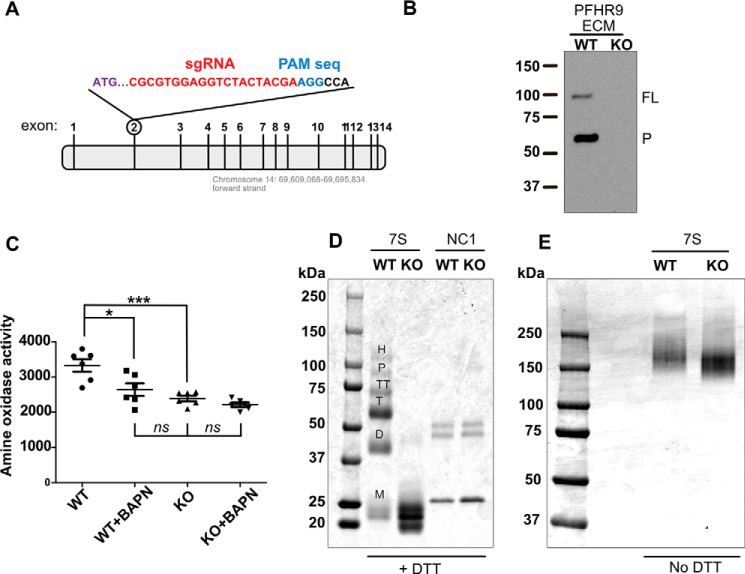
Figure 3.
CRISPR/Cas9-mediated deletion of LOXL2 in PFHR9 cells yields 7S dodecamers devoid of lysyl-derived crosslinks. A, representation of the Loxl2 gene structure and the region of exon 2 targeted by the sgRNA sequences. B, immunoblotting analysis using an anti-LOXL2 antibody of ECM proteins extracted with urea from wild-type (WT) and LOXL2-null (KO) PFHR9 cells. A representative experiment from a total of three independent experiments is shown. Full-length (FL) and processed (P) forms of LOXL2 are indicated. C, the graph represents the fluorescence units generated by Amplex Red assay measuring the amine oxidase activity from urea-soluble matrix proteins from LOXL2-KO and WT PFHR9 cells in the presence or absence of BAPN. Error bars represent S.D. from five independent experiments. Data were analyzed by Student's t test. Statistical significance is shown above the bars. ***, p value ≤ 0.001; *, p value ≤ 0.05; ns, not significant. D, SDS-PAGE analyses of 7S dodecamer (7S) and NC1 hexamer (NC1) purified from LOXL2-KO and WT PFHR9 matrices. Proteins were reduced with 50 mm DTT. M, D, T, TT, P, and H indicate the electrophoretic mobility of monomer, dimer, trimer, tetramer, pentamer, and hexamer 7S subunits, respectively. E, SDS-PAGE analyses under nonreducing conditions of the same 7S dodecamers purified from WT and KO PFHR9 cell matrices.