Figure 6.
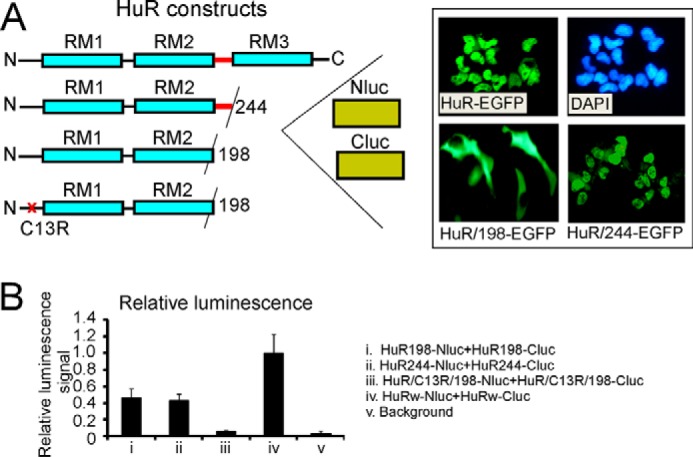
Assessment of HuR oligomerization domains in split firefly luciferase assay using reporter constructs with truncated and mutated HuR. A, schematic illustration of truncated and mutated HuR constructs used in split firefly luciferase assays (see “Experimental procedures”). The inset illustrates subcellular distribution of HuR-EGFP, HuR/244-EGFP (without RRM3 (RM3)), and HuR/198-EGFP (without hinge and without RRM3) constructs. Note that HuR proteins with deleted hinge regions lack the HuR nuclear localization signal and reside in the cytoplasmic fraction. The HuR proteins with only RRM3 domains deleted have nuclear/cytoplasmic distribution similar to that of full-length HuR. The subcellular distribution of the above constructs is in agreement with previous data (28). B, graph representing averaged relative luminescence signals from cells coexpressing dox-inducible HuR/198-Nluc and HuR/198-Cluc (i), HuR/244-Nluc and HuR/244-Cluc (ii), HuR/C13R/198-Nluc and HuR/C13R/198-Cluc (iii), and HuRw-Nluc and HuRw-Cluc (iv) constructs. Note that averaged luminescence signals from HuR/198-Nluc and HuR/198-Cluc (i), HuR/244-Nluc and HuR/244-Cluc (ii), and HuRw-Nluc and HuRw-Cluc (iv) cell lines significantly exceed background luminescence signal (v). The error bars represent S.D. for each group.
