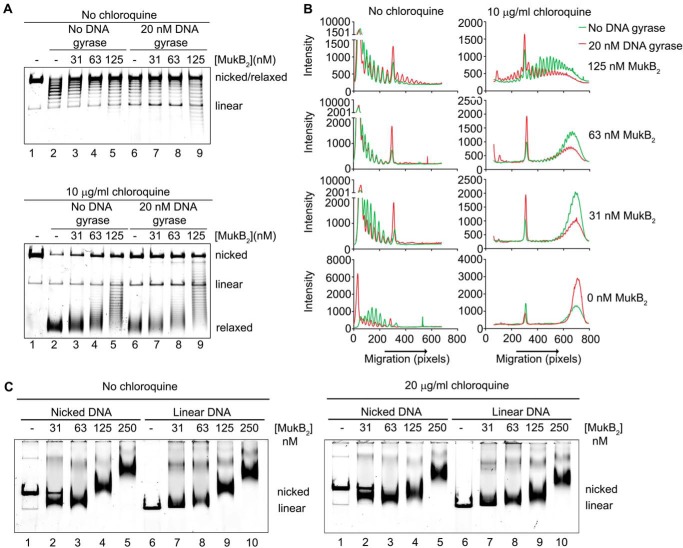
Figure 2.
Loops formed by MukB in the DNA are topologically isolated. A, DNA loops stabilized by MukB can be supercoiled by DNA gyrase. MukB was incubated with the nicked DNA substrate, 2 mm ATP, and 20 nm DNA gyrase for 30 min at 37 °C. Novobiocin was added to 10 μm and the incubation continued for 5 min. Bacteriophage T4 DNA ligase was then added and the incubation continued for 30 min. The products were deproteinized before analysis by electrophoresis through agarose gels either in the absence (top panel) or presence (bottom panel) of 10 μg/ml chloroquine in both the gel and the running buffer. Relaxed, the nicked DNA sealed by DNA ligase to give a closed DNA ring that does not contain supercoils. B, densitometric lane traces of the gels shown in A. C, MukB stabilizes topologically isolated loops in linear DNA. MukB was incubated with either nicked DNA (lanes 1–5) or linear DNA (blunt-end cut, lanes 6–10) for 5 min at 37 °C. Protein-DNA complexes were then analyzed as in Fig. 1D above.