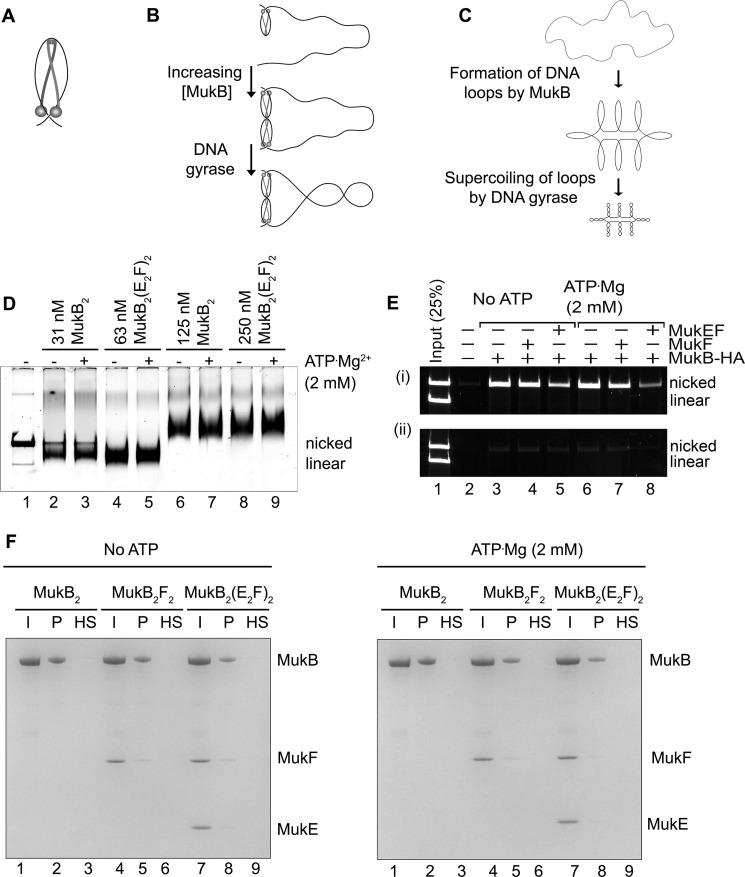
Figure 6.
Model for MukB condensation of DNA. A, MukB protects a negative supercoil when bound to DNA. We propose that DNA binding by both the head domain and hinge domain is necessary for efficient negative supercoiling. The disposition of the DNA on the MukB hinge should not be taken literally. Whereas the amino acid residues mutated in the KE,RE variant are on the top of the hinge, we do not know precisely how DNA is bound to the hinge. One presumes that the DNA binds in such a manner that it does not interfere with MukB oligomerization via hinge–hinge interactions. B, MukB can form stable, topologically isolated loops in the DNA via hinge–hinge interactions between dimers. C, progressive DNA condensation by MukB is driven by the formation of topological loops in the DNA that can be further negatively supercoiled by DNA gyrase. D, effect of MukEF and ATP on MukB-DNA complexes. The indicated concentrations of MukB or the MukBEF complex in the presence or absence of ATP were incubated for 5 min at 37 °C. Protein-DNA complexes were then analyzed as described in the legend to Fig. 1A. Note that twice the concentration of MukBEF was required to observe similar DNA-binding activity compared with MukB alone. E, MukB does not trap DNA topologically. The indicated concentrations of MukB-HA, MukF, and MukEF, either in the presence or absence of ATP, were incubated with an equal mixture of nicked and linear DNA for 5 min at 37 °C. Panel i, anti-HA antibody attached to magnetic beads was then added and immediately thereafter NaCl was added to 0.5 m, the beads were washed in 0.75 m NaCl after pulldown, and the bound DNA was released by proteinase K digestion and analyzed by agarose gel electrophoresis. Panel ii shows the same experiment except that 0.5 m NaCl was added immediately before addition of the antibody beads. F, MukB does not trap DNA topologically. Tailed, nicked plasmid DNA attached to magnetic beads was incubated with the indicated concentrations of MukB, MukF, and MukEF either in the presence or absence of ATP in standard reaction buffer (low salt) for 5 min at 37 °C. The beads were then pulled down and resuspended in buffer containing 0.5 m NaCl. The beads were pulled down again, and the samples were resuspended in 1× SDS-PAGE loading dye. After heating to 95 °C, the proteins released were analyzed by SDS-PAGE. I, input; P, DNA bound to the beads after the first pulldown in low salt; HS, DNA bound to the beads after washing the beads in high salt.