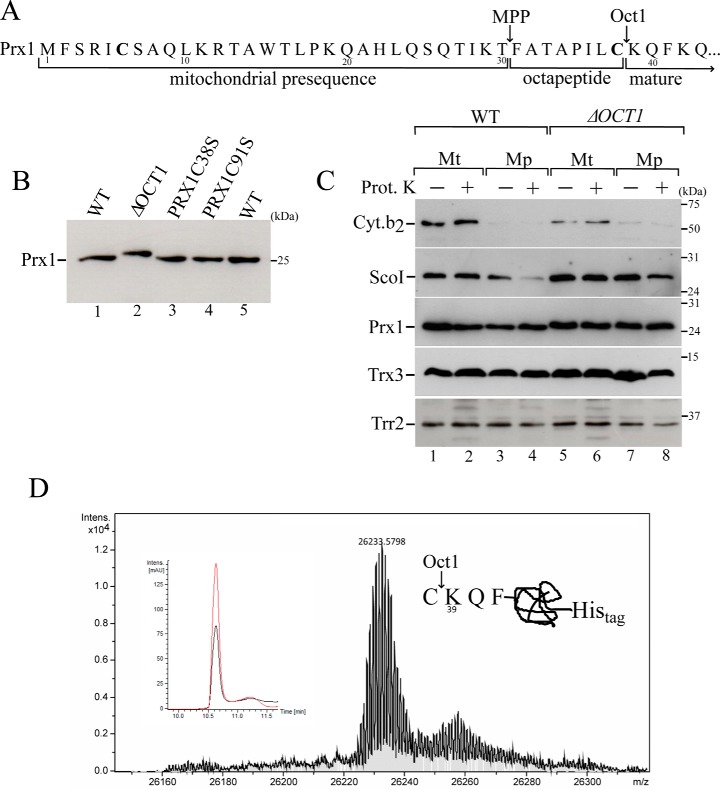
Figure 2.
Mitochondrial processing of Prx1 by Oct1 protease. A, scheme of the Prx1 N-terminal amino acid sequence. Arrows indicate the MPP and Oct1p cleavage sites based on a global mass spectrometric analysis (27). Cysteines present in the Prx1 presequence are highlighted in bold. B, total cell-free extracts from the wild type (WT, BY4741), OCT1 null mutant (ΔOCT1), and PRX1 null mutant expressing cysteine mutant versions of PRX1 (PRX1C38S and PRX1C91S) were analyzed by Western blotting. C, intramitochondrial localization of Prx1 in the wild-type and ΔOCT1 mitochondria (Mt). Mp, mitoplasts. Mitochondria isolated from cells grown on YPGal were processed according to legend of Fig. 1B. D, mass spectrometry analysis of C-terminal His-tagged Prx1 present in the matrix fraction. C-terminal His-tagged Prx1 expressed in the ΔPRX1 strain was purified by nickel affinity chromatography. The obtained mass was 26,233.57, which was consistent with the incorporation of two oxygen atoms to the Prx1 polypeptide. The inset depicts the corresponding reverse phase chromatogram. The red and black lines correspond to UV chromatogram at 280 and 254 nm, respectively. The scheme at the right side of the mass spectrum represents the cleavage site. These results are representative of at least two independent biological replicates, each one in two technical replicates. See Table 2 for strains and genotypes.