Abstract
The culture fluid of HBV replicating cells contains a mixture of viral particles with different structural and genetic components, including enveloped infectious virions, genome-free virion, envelope-only subviral particles, and nonenveloped naked capsids. Based on their different physical and chemical properties, the enveloped and nonenveloped particles can be separated by the native agarose gel electrophoresis and transferred onto a positively charged microporous membrane, then the virus particle-associated protein components and nucleic acid content can be detected by antibody-based enzyme immunoassay (EIA) and hybridization, respectively. Such convenient experimental procedure is called HBV particle assay and described in detail in this chapter. The particle gel assay can be used to study viral and host regulations of HBV virus morphogenesis and egress, and for antiviral assessment of HBV inhibitors as well.
Keywords: HBV particles, Particle gel assay
1 Introduction
Hepatitis B virus (HBV) is an enveloped DNA virus belonging to the Hepadnaviridae family [1]. The infectious HBV virion is comprised of a single copy of approximately 3.2 kb genomic relaxed circular DNA (rcDNA) within an icosahedral capsid (core) which is enveloped by a lipid bilayer shell studded with complexes of viral glycoproteins including “large” (L), “medium” (M), and “small” (S) surface proteins [2, 3]. The three HBV surface proteins are all synthesized from the same open reading frame of the viral genome by using different starting codons. They share the common S domain at the C-terminus. The M and L proteins contain S plus pre-S2 and S plus pre-S2 and pre-S1 domains, respectively. S protein is the major viral surface protein. An HBV virion contains approximately 100 copies of S protein for every five copies of M and one copy of L protein [2].
Besides infectious virion particles, two additional forms of HBV particles are routinely found in hepatitis B patient blood, which are, the envelope-only particles (also called subviral particles) and the DNA-free empty virions (enveloped empty capsid) [4]. Several groups have demonstrated that HBV virion and subviral particles egress the host cell by different routes. The budding of 42-nm-diameter virion particles takes place at intracellular membranes through the multivesicular bodies (MVB) pathway, a process dependent on the endosomal sorting complex required for transport (ESCRT) system that consists of the ESCRT-0, -I, and -III complexes together with the Vps4 ATPase and other associated proteins [5–7]. The production of virions is always accompanied by the formation of subviral particles, which are 20-nm-diameter spherical and filamentous particles consisting only of lipid and HBV envelope proteins, predominantly S protein [8, 9]. HBV subviral particles are assembled on the ER membrane, and secreted through the constitutive ER-Golgi secretory pathway [10]. Normally, a HBV infected cell secretes excess amounts of empty virions over infectious virions (usually ≥100-fold) [11], and hundreds to thousands of subviral particles for every virion [12]. Both DNA-free enveloped particles are known as hepatitis B surface antigen (HBsAg) and likely assumed to serve as immunological decoy to mask the bona fide infectious virions from host immune surveillance and attack [11]. In addition to enveloped particles, nonenveloped capsids (naked capsids) have also been found to be noncytolytically released into the culture fluid of HBV-replicating cells usually at a large excess to virions, though they are not detected in the blood of infected patients [4, 13–15]. Although a number of host proteins have been implicated to regulate naked capsid egress, the secretion pathway for naked capsids and their biological significance remains elusive [16–18].
In order to study the morphogenesis and secretion of hepadnavirus particles, Lenhoff and Summers invented a native agarose gel based particle assay for duck hepatitis B virus (DHBV) [19], which was later adapted for the study of HBV and woodchuck hepatitis virus (WHV) [4, 13, 14, 18, 20]. The principle of particle gel assay is built upon the different migration rate of HBV particles in native agarose gel electrophoresis due to their different composition, particle size, and electric charge, which permits separation of enveloped particles from naked capsids. The virus particles are then transferred to a membrane for enzyme immunoassay (EIA) with antibodies against HBsAg or core. Furthermore, viral DNA within the nucleocapsids can be released in situ and detected by DNA hybridization. Currently, quantitative PCR (qPCR) is widely used to measure the amount of infectious virions (or virus genome equivalent, or vge) secreted from HBV-replicating cells, especially in antiviral assessment assays and preparation of HBV inoculum for infection assays. However, because it does not discriminate between virion particles and genome-containing secreted naked capsids, the qPCR will overestimate the levels of infectious vge. Therefore, the particle gel assay also provides a convenient and reliable method for measuring the ratio between virion DNA and capsid DNA [21]. We describe herein a protocol for HBV particle gel assay in detail.
2 Materials
2.1 Precipitation of HBV Particle from Cell Cultured Media
35 % PEG8000 in 1.5 M NaCl.
TNE buffer: 10 mM Tris–HCl (pH 7.4), 150 mM NaCl, and 1 mM EDTA.
2.2 Agarose Gel Electrophoresis
10× DNA gel loading buffer: 10 mM EDTA (pH 8.0), 50 % (V/V) glycerol, 0.25 % (W/V) bromophenol blue.
1× TAE buffer: 0.04 M tris base, 0.04 M glacial acetic acid, 1 mM EDTA (pH 8.2–8.4). Prepare 30× stock solution, store at room temperature.
Agarose (molecular biology grade).
2.3 Particle Gel Transfer
TNE buffer: 10 mM Tris–HCl, pH 7.4, 150 mM NaCl and 1 mM EDTA.
Protran BA85 Nitrocellulose (NC) Blotting Membrane, 0.45 μm (GE Healthcare).
3MM Chromatography (Chr) paper (GE Healthcare).
2.4 Enzyme Immunoassay
Polyclonal rabbit anti-HBV core antigen (Dako).
Monoclonal mouse anti-HBV surface antigen (Dako).
IRDye 800CW goat anti-rabbit IgG (H + L) (LI-COR Biosciences).
IRDye 800CW goat anti-mouse IgG (H + L) (LI-COR Biosciences).
WesternBreeze Blocker/Diluent (Part A and B) (Life Technologies).
WesternBreeze Wash Solution (Life Technologies).
Phosphate buffered saline (PBS).
Formaldehyde.
Methanol.
Odyssey Imager (LI-COR Biosciences).
2.5 HBV DNA Riboprobe
pGEM-3Z vector (Promega) with an insertion of genome-length HBV DNA fragment (EcoRI linearized), in which the transcription of HBV sense RNA is under the control of prokaryotic SP6 promoter.
800 Ci/mmol [α-32P] UTP.
Riboprobe in vitro transcription system-Sp6 (Promega).
5 M NH4OAc.
10 mg/mL yeast RNA (Ambion).
Isopropanol.
Formamide.
2.6 Southern Blotting and DNA Hybridization
Denaturing buffer: 0.5 M NaOH, 1.5 M NaCl.
Neutralization buffer: 1.5 M NaCl, 1 M Tris–HCl (pH 7.4).
UV cross-linker.
EKONO hybridization buffer (G-Biosciences).
Hybridization wash buffer: 0.1 % SDS, 0.1× SSC.
Phosphorimager system (GE Healthcare).
3 Methods
3.1 Cell Cultures
Culture HBV-producing cells, such as transiently transfected HepG2 or Huh7 cells, or stable cell lines like HepG2.2.15 or tetracycline-inducible (tet-off) HepAD38 or HepDE19 cells, in collagen coated cell culture flask or plate at approximately 100 % confluence with appropriate culture medium.
24–48 h post transfection or seeding, change the culture medium with fresh medium. For tetracycline inducible cell lines, remove tetracycline from culture medium to induce HBV replication.
Change and harvest the culture medium at designed time interval for specific experiment. Clarify the harvested supernatant by centrifugation at 1000 × g for 10 min at 4 °C. Transfer the supernatant to a new tube, store at −80 °C if not used immediately.
3.2 HBV Particle Precipitation from Cell Cultured Media
Transfer 1 mL of harvested cell culture supernatant to a 1.7 mL Eppendorf tube. Add 400 μL of 35 % PEG8000 to reach final concentration of 10 %, and rotate the tube on a rotisserie for at least 2 h at 4 °C.
Centrifuge at 10,000 × g for 5 min at 4 °C to precipitate particles (see Note 1).
Discard the supernatant completely. Submerge the pellet in 20 μL TNE buffer with 1× DNA loading buffer to dissolve the virus particles. Let the tube sit at 4 °C overnight (see Note 2).
3.3 Agarose Gel Electrophoresis
Prepare 150 mL of 1 % agarose gel by mixing 1.5 g agarose, 5 mL of 30× TAE buffer, and 145 mL of distilled water in a microwavable glass bottle. Microwave the mixture until the agarose is thoroughly melted and dissolved in the solution. Let it cool down to about 50 °C at room temperature. Pour the liquid gel into the gel box with an appropriate comb inserted. Let the gel solidify at room temperature for at least 2 h before removing the comb.
Prepare 600 mL of gel running buffer by diluting 20 mL of 30× TAE buffer in 580 mL of deionized water.
Assemble gel running apparatus by correctly positioning the gel box, adding running buffer into the chamber.
Pipette up and down very gently with an open mouth tip to mix the dissolved virus particle samples, and load each sample into a separate well (see Note 3). Connect the electrophoresis unit to a power supply and run the gel at 23 V constantly overnight.
3.4 Particle Gel Transfer
Disconnect the gel running unit and take out the gel carefully. Trim the gel to appropriate size.
Prepare the NC membrane with the same size of the gel. Submerge the membrane in a tray with deionized water for 5 min at room temperature. Replace the deionized water with TNE buffer and continue to agitate for 15 min.
To transfer particles from the gel to the NC membrane, set up the transfer cassette as following: Layer two sheets of 8″ × 11″ Whatman 3MM Chr paper in the gel transfer tray and pour TNE buffer on top to completely wet the Chr papers. Smooth out the bubbles in between the Chr papers and the tray gently with a plastic roller, and get rid of the excess amount of the buffer. Place the gel on top of the Chr papers with the loading well side facing down, make sure there is no air bubble between the papers and the gel. Place the presoaked NC membrane on top of the gel, and use a roller to get rid of the air bubbles between the gel and the membrane. Layer two sheets of TNE buffer pre- wet Chr papers that are cut to the gel size on top of the membrane, and use a roller to remove bubbles in between. Place a stack of dry paper towels (about 4 in.) precut to the gel size on top of the Chr papers. Seal the whole transfer tray with a big piece of plastic wrap to prevent buffer evaporation during transfer. Put some weight (approx. 500 g such as metal plate, casserole dish) on top of the transfer apparatus (Fig. 1). Let the assembled transfer apparatus sit still on a flat surface and transfer for 24 h (see Note 4).
After gel transfer, take out the membrane with the gel still attached, and mark the position of the loading wells on the membrane by a pencil (see Note 5). Wrap up the gel and paper towels by plastic wrap and discard.
Fig. 1.
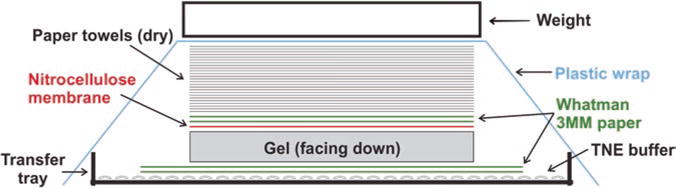
Assembly of HBV particle gel transfer apparatus. See Subheading 3.3 for details
3.5 Enzyme Immunoassay
Prepare WesternBreeze blocking buffer with the ratio of H2O–part A–part B as 7:2:1 (see Note 6).
Soak membrane in PBS buffer containing 2.5 % formaldehyde at room temperature for 10 min (see Note 7).
Rinse the membrane with deionized water.
Fix the membrane with 50 % methanol at room temperature for 30 min.
Wash the membrane three times with deionized water.
Incubate the membrane in blocking buffer for 1 h at room temperature.
Dilute anti-hepatitis B virus core antigen or anti-hepatitis B virus surface antigen in blocking buffer (see Note 6). Incubate with blot for 1–3 h.
Wash the membrane four times for 10 min in WesternBreeze wash solution (see Note 6).
Dilute IRDye 800CW secondary antibody in blocking buffer (1:10,000). Incubate the membrane for 1 h.
Wash four times for 10 min in WesternBreeze wash solution.
Dry the membrane by using a filter paper, then scan the membrane with Odyssey imager system.
The typical image of HBV particle surface and core EIA is shown in Fig. 2a and b, respectively.
Fig. 2.
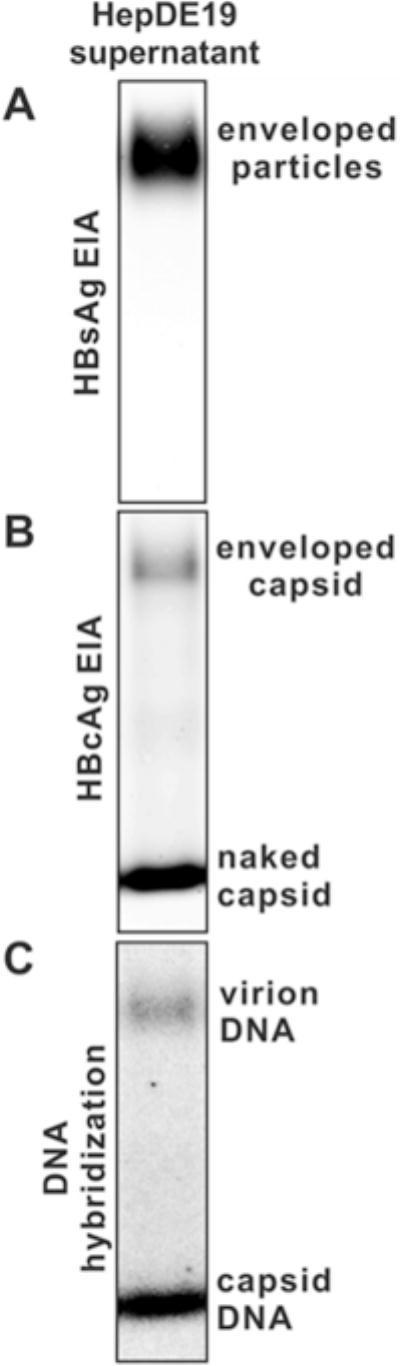
Detection of HBV particles released into cell culture fluid by particle gel assay. HBV virus particles in the supernatant of HepDE19 cells cultured in tetracycline- free medium were precipitated down and analyzed by particle gel assay described in this chapter. (a) HBsAg EIA of enveloped HBV particles, including virions and subviral particles. (b) HBcAg EIA of Virion-containing capsids and nonenveloped naked capsids. (c) In situ hybridization of HBV DNA in virions and naked capsids. Reproduced from [14] with permission from American Society for Microbiology
3.6 Release of Particle- Associated DNA on Membrane
Submerge the particle bonded membrane in a tray containing denaturing buffer, and agitate gently for 15 min at room temperature (see Note 8).
Rinse the membrane three times with distilled water.
Submerge the membrane in a tray of neutralization buffer, and agitate gently for 5 min at room temperature.
Dry the membrane briefly in air at room temperature.
Cross-link the NC membrane in a UV cross-linker with UV energy dosage at 120 mJ/cm2. The membrane can be directly subject to hybridization, or sandwiched between two sheets of filter papers and stored at −20 °C.
3.7 Riboprobe Preparation for HBV DNA Detection (See Note 9)
Prepare rNTP solution by mixing 50 μL of 10 mM ATP, 50 μL of each 10 mM rGTP, 10 mM rCTP, and 0.2 mM UTP.
Add 4 μL of 5× reaction buffer, 4 μL of NTP solution from the previous step, 2 μL of 100 mM DTT, 0.25 μg of Sal I-linearized pGEM-HBV DNA template, 1 μL of RNasin, and 1 μL of Sp6 polymerase into a nuclease free tube. Adjust the volume to 14 μL with nuclease free water before adding 6 μL of [α-32P] UTP into the mixture. Incubate the reaction at 37 °C for 1 h.
To digest the DNA template, add 1 μL of RNase-free RQ1 DNase to the reaction and incubate at 37 °C for 15 min.
Add 15 μL of 5 M NH4OAc to stop the reaction. To precipitate the RNA probe, add 113 μL of nuclease free water, 2 μL of yeast RNA, and 150 μL of isopropanol, gently mix and incubate at room temperature for 10 min.
Centrifuge the mixture at 12,000 × g for 30 min at 4 °C, and discard the supernatant carefully without disturbing the pellet. Dissolve the probe in 400 μL of deionized formamide, followed by measurement of the counts per minute (CPM) of acid-insoluble 32P with scintillation counter (PerkinElmer). Store the probes at −20 °C.
3.8 DNA Hybridization
Place the cross-linked membrane in a hybridization tube with the DNA-binding side facing the center of the tube. Add 5 mL of EKONO hybridization buffer, and pre-hybridize the membrane by rotating the hybridization tube at 65 °C for 1 h in a hybridization oven (see Note 10).
Replace the pre-hybridization buffer with 5 mL of fresh hybridization buffer, and add HBV riboprobes with 1 × 107 CPM. Rotate the hybridization tube at 65 °C overnight.
Discard the hybridization solution on the following day, and wash the hybridization membrane with approximately half a tube of wash buffer. Rotate at 65 °C for 30 min.
Discard the previous wash buffer, and replace with half a tube of fresh wash buffer. Continue to wash at 65 °C for 1 h.
After the second wash, take out the membrane and dry it with paper towels. Seal the membrane with plastic wrap. Expose the membrane to the phosphorimager screen in a closed cassette for overnight.
Scan the phosphorimager screen with Typhoon phosphorimager system (GE Healthcare) for quantitation of signal intensity of virion and capsid DNA (see Note 11). A typical result of HBV particle DNA hybridization is shown in Fig. 2c.
Acknowledgments
We thank Dr. Jesse Summers for inventing the hepadnavirus particle gel assay. This work was supported by NIH grants (R01AI094474, R01AI110762, and R21AI103838).
Footnotes
The precipitated particles will be difficult to dissolve if the centrifugation speed is higher than 10,000 × g or the time of centrifugation is more than 5 min.
Do not try to dissolve the pellet by pipetting up and down, which may physically damage the particles. The pellet will be slowly dissolved in TNE buffer at 4 °C.
There may be a small piece of gel-like pellet that cannot be dissolved at all. Do not load the undissolved pellet into the gel well.
When assembling the transfer apparatus, avoid possible short circuiting of capillary liquid flow. Use Parafilm strips to seal the edges of the gel.
After transfer, the gel becomes thinner. Use a pencil tip to penetrate the loading wells and mark their positions on the membrane. This procedure is used to label the particle binding side of the membrane and the gel lanes.
Each lab may use their preferred Western blot blocking buffer, antibody dilution buffer, and washing buffer.
Formaldehyde is used to disinfect HBV virions and cross-link the proteins to membrane.
The same particle gel membrane can be subjected to DNA hybridization after EIA, or prepare a duplicate for DNA detection only.
Radiolabeled DNA probes can also be used for hybridization.
If other types of hybridization buffers are used, follow the pre- hybridization and hybridization conditions specified by the manufacturers.
When preparing HBV virus stocks from cell cultures for infection assays, the ratio of virion DNA–capsid DNA is needed to calculate the actual infectious vge of the inoculum [21].
References
- 1.Summers J, O’Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci U S A. 1975;72(11):4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block TM, Guo H, Guo JT. Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis. 2007;11(4):685–706. doi: 10.1016/j.cld.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64(1):51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, Hafenstein S, Hu J. Secretion of genome-free hepatitis B virus—single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 2011;7(9):e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert C, Doring T, Prange R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol. 2007;81(17):9050–9060. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A. 2007;104(24):10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prange R. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol. 2012;201(4):449–461. doi: 10.1007/s00430-012-0267-9. [DOI] [PubMed] [Google Scholar]
- 8.Bruss V. Hepatitis B virus morphogenesis. World J Gastroenterol. 2007;13(1):65–73. doi: 10.3748/wjg.v13.i1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci U S A. 2005;102(41):14783–14788. doi: 10.1073/pnas.0505062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patzer EJ, Nakamura GR, Simonsen CC, Levinson AD, Brands R. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J Virol. 1986;58(3):884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luckenbaugh L, Kitrinos KM, Delaney WE, Hu J. Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J Viral Hepat. 2015;22(6):561–570. doi: 10.1111/jvh.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlich WH, Glebe D, Schuttler CG. Deficiencies in the standardization and sensitivity of diagnostic tests for hepatitis B virus. J Viral Hepat. 2007;14(Suppl 1):16–21. doi: 10.1111/j.1365-2893.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81(22):12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H, Pan X, Mao R, Zhang X, Wang L, Lu X, Chang J, Guo JT, Passic S, Krebs FC, Wigdahl B, Warren TK, Retterer CJ, Bavari S, Xu X, Cuconati A, Block TM. Alkylated porphyrins have broad antiviral activity against hepadnaviruses, flaviviruses, filoviruses, and arenaviruses. Antimicrob Agents Chemother. 2011;55(2):478–486. doi: 10.1128/AAC.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Possehl C, Repp R, Heermann KH, Korec E, Uy A, Gerlich WH. Absence of free core antigen in anti-HBc negative viremic hepatitis B carriers. Arch Virol Suppl. 1992;4:39–41. doi: 10.1007/978-3-7091-5633-9_8. [DOI] [PubMed] [Google Scholar]
- 16.Bardens A, Doring T, Stieler J, Prange R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT- independent manner. Cell Microbiol. 2011;13(4):602–619. doi: 10.1111/j.1462-5822.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doring T, Prange R. Rab33B and its autophagic Atg5/12/16L1 effector assist in hepatitis B virus naked capsid formation and release. Cell Microbiol. 2015;17(5):747–764. doi: 10.1111/cmi.12398. [DOI] [PubMed] [Google Scholar]
- 18.Chou SF, Tsai ML, Huang JY, Chang YS, Shih C. The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PLoS Pathog. 2015;11(10):e1005123. doi: 10.1371/journal.ppat.1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68(7):4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo JT, Guo H. The interferon-inducible protein tetherin inhibits hepatitis B virus virion secretion. J Virol. 2015;89(18):9200–9212. doi: 10.1128/JVI.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan R, Zhang Y, Cai D, Liu Y, Cuconati A, Guo H. Spinoculation enhances HBV infection in NTCP-reconstituted hepatocytes. PLoS One. 2015;10(6):e0129889. doi: 10.1371/journal.pone.0129889. [DOI] [PMC free article] [PubMed] [Google Scholar]


